Specific primer pair and kit for detecting methylation of septin9 gene according to high-resolution melting curve
A technology of specific primer pairs and kits, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve problems such as difficulty in achieving concentration, and achieve simple operation, good specificity and low cost. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Embodiment 1, design and preparation of primers and preparation of standard plasmids
[0047] 1. Design and preparation of primers
[0048] Multiple primer pairs were designed based on the CpG island of septin9 gene, and the effects of multiple primer pairs were verified through preliminary experiments, and 3 primer pairs were obtained through preliminary screening.
[0049] Primer pair A consists of primer F1 and primer R1.
[0050] F1 (SEQ ID NO: 1): 5'-GTTAGTGCGAGATAGGGAGGTC-3';
[0051] R1 (SEQ ID NO: 2): 5'-CAATACAAAACTAACCGCCG-3'.
[0052] Primer pair B consists of primer F2 and primer R2.
[0053] F2: 5'-AGAAATTTTAGGATTTGGGCG-3';
[0054] R2: 5'-GACGTTTCCCACCTACTTAAACG-3'.
[0055] Primer pair C consists of primer F3 and primer R3.
[0056] F3: 5'-GTGTTGGGTTATAAGACGTTAGAATC-3';
[0057] R3: 5'-TAACAAAACAATAAACACCTCGAA-3'.
[0058] The above primers were synthesized separately.
[0059] 2. Preparation of standard quality particles
[0060] Preparation of...
Embodiment 2
[0062] Example 2. Establishment of a method for detecting methylation by methylation-specific PCR combined with HRM curve analysis
[0063] 1. Bisulfite treatment
[0064] Take 20 μl of the sample solution and perform bisulfite treatment to obtain 10 μl of the product solution, which is the template solution. The kit used for bisulfite treatment is: EZ-96 DNA Methylation-Gold MagPrep (ZYMO, product number D5042); operate according to the kit instructions.
[0065] 2. Perform PCR amplification and high-resolution melting curve analysis
[0066] The reaction system is shown in Table 1.
[0067] The instrument used for the reaction is 480II (Roche).
[0068] The amplification reaction program is shown in Table 2.
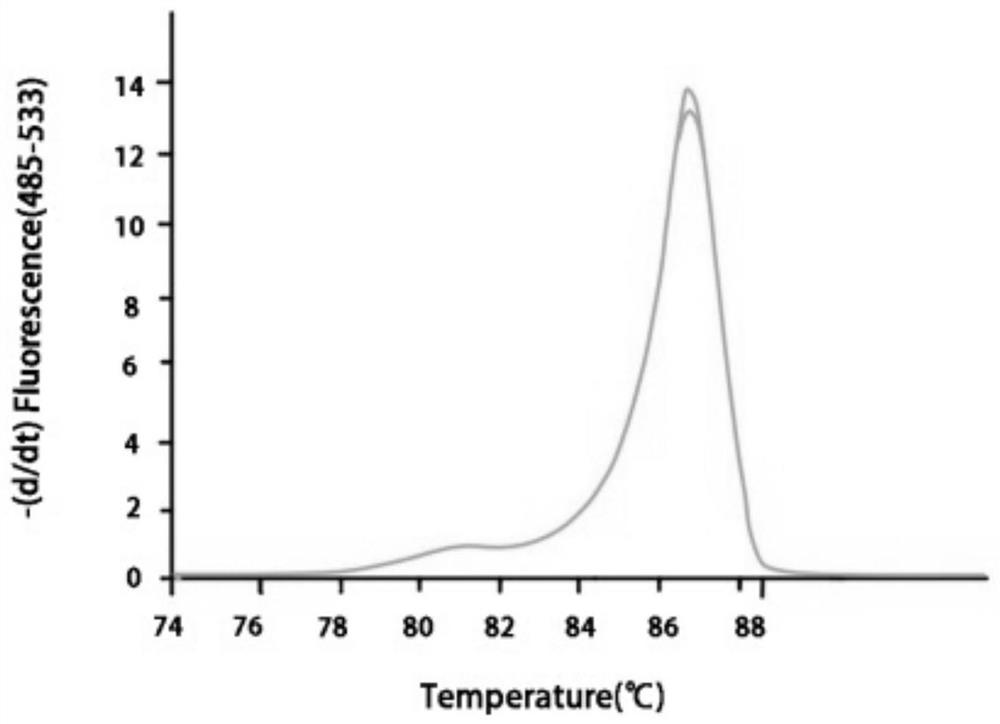
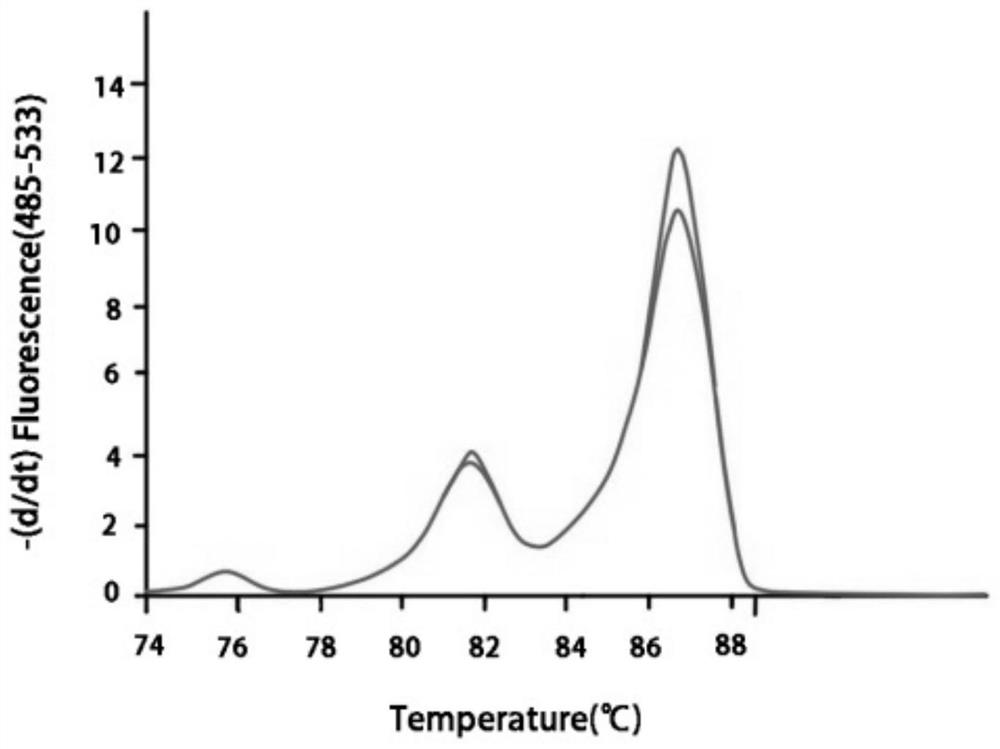
[0069] After amplification, high-resolution melting (high-resolution melting, HRM) curve analysis was performed. The parameters of the HRM curve analysis were set as follows: 65°C for 1s, then increased from 65°C to 95°C at a rate of 0.02°C / s (25 fluorescence co...
Embodiment 3
[0082] Embodiment 3, sensitivity detection
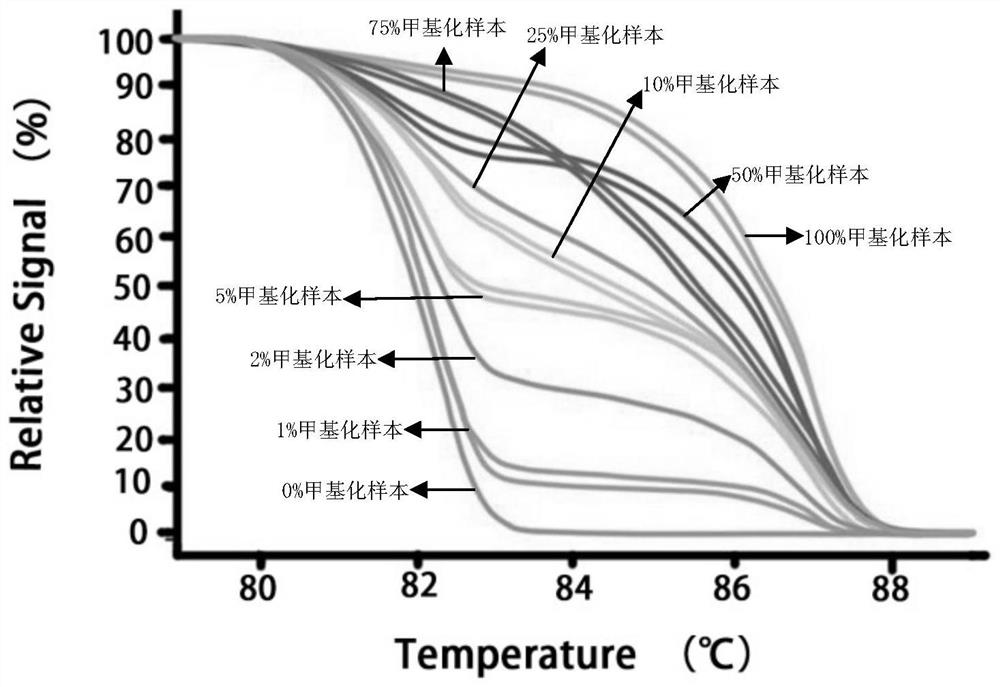
[0083] Prepare sample solution, also known as 100% methylated sample, with methylated plasmid and TE buffer. A sample solution was prepared by using methylated plasmids, unmethylated plasmids and TE buffer (the mass percentage of methylated plasmids to the total plasmids was 75%), also known as 75% methylated samples. A sample solution was prepared with methylated plasmids, unmethylated plasmids and TE buffer (the mass percentage of methylated plasmids in the total plasmids was 50%), also known as 50% methylated samples. A sample solution was prepared by using methylated plasmids, unmethylated plasmids and TE buffer (the mass percentage of methylated plasmids to the total plasmids was 25%), also known as 25% methylated samples. A sample solution was prepared by using methylated plasmids, unmethylated plasmids and TE buffer (the mass percentage of methylated plasmids in the total plasmids was 10%), also known as 10% methylated sampl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com