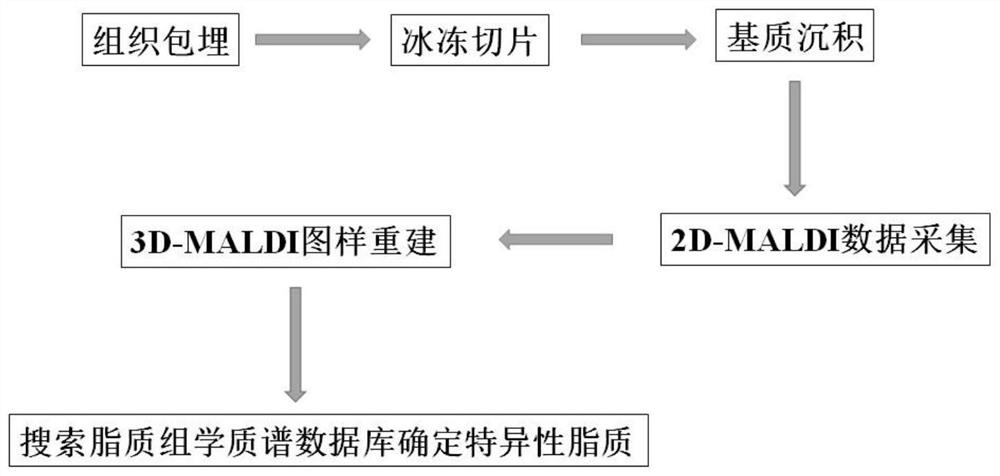
In-vivo lipid three-dimensional mass spectrum imaging method based on integral zebra fish model
A mass spectrometry imaging, zebrafish technology, applied in the field of mass spectrometry detection, can solve problems such as spleen enlargement, mental retardation, dystonia, etc., and achieve the effect of high chemical specificity and high integrity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1 Acquisition of Whole Zebrafish Mass Spectrometry Imaging Data
[0049] Sample preparation for frozen sections: NPC1 - / - After the adult zebrafish in group and WT group were euthanized, the whole zebrafish was embedded with embedding agent, and placed in a -80°C refrigerator until it solidified; after that, frozen section samples were prepared and placed in a cryostat at -20°C Take a tissue section sample with a thickness of 14 μm every 56 μm and adhere to the ITO plate, NPC1 - / - 20 slices were taken from the control group and the WT group respectively. Place the ITO plate with the tissue slice attached in the vacuum drying oven for 30 min before depositing the NEDC matrix, and each plate needs about 6 mL.
[0050] Matrix deposition instrument: matrix deposition instrument (Imageprep, Bruker Company). Deposition conditions: 35% of spray power; modulation 20%; deposition time 1.0s; incubation time 15s; drying time 60s.
[0051] Mass Spectrometry Instrument: ...
Embodiment 2 3
[0052] Embodiment 2 Construction of three-dimensional MALDI mass spectrometry image
[0053] The mass spectrometry imaging data was imported into SCiLS Lab 2016b (Bremen, Germany) Premium 3D for processing. By adjusting the x and y axes, 20 tissue slices could be accurately stacked according to the original biological appearance, making it approximately represent the whole animal model. Normalization was performed with the total ion current method, the m / z interval was set to ±0.1Da, and all MALDI mass spectrometry images were processed using a weak denoising method, and then the mass spectrometry images of lipids in the original position of the tissue were constructed.
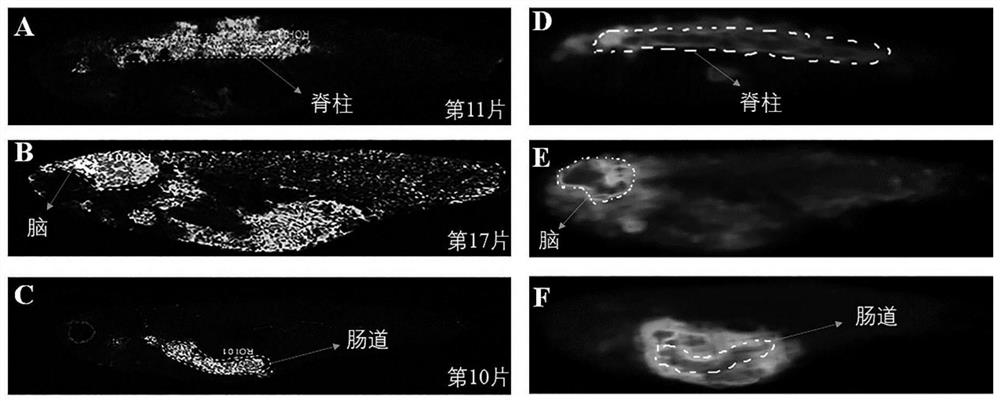
[0054] Such as figure 2 Shown: figure 2 A. figure 2 B. figure 2 C are two-dimensional MALDI mass spectrometry images distributed in the spine (m / z 452.324), brain (m / z 790.854) and intestinal tract (m / z 867.534). The largest sections of the three organs are located in the 11th slice , on the 17th and ...
Embodiment 3
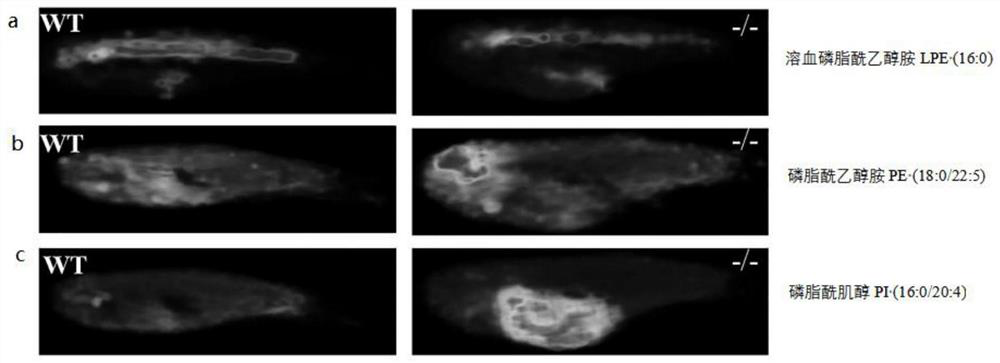
[0055] Example 3 Screening of differential lipids in the zebrafish NPC1 disease model
[0056] Embedded NPC1 after euthanasia - / - The whole zebrafish of the group and the WT group, after obtaining the frozen section tissue samples and carrying out matrix deposition, according to the method of embodiment 1, respectively to NPC1 - / - MALDI mass spectrometry imaging was performed on all sliced samples of the WT group and the WT group, and then the three-dimensional MALDI patterns were reconstructed according to the method in Example 2. Fifteen kinds of differential lipid m / z were screened, and the detected differential m / z values were entered into the lipidomics mass spectrometry database Lipidsearch4.0 for searching, and compared with the secondary fragment information to determine the differential m / z The molecular formula corresponding to the z value and the name of the lipid corresponding to the molecular formula. The search conditions are: the molecular weight deviation...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com