DNA framework-coated G-quadruplex structure as well as preparation method and application thereof
A framework structure and DNA strand technology, applied in the field of DNA framework-wrapped G-quadruplex structure and its preparation, can solve the problems of high background signal, poor permeability and the like, and achieve low cost, simple preparation method and high cell permeability effect on the ability to hybridize to the target
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] The 8 sequences used to assemble the DNA framework were first quantified to 100 μM, wherein the 8 sequences included the 6 DNA sequences C1-C6 required for the assembly of the framework structure, and the Cy3 sequence C7a required to form the G-quadruplex probe and Cy5 sequence C7b, the linear G-quadruplex probe targets miR-122a, as shown in Table 1 below:
[0030] Table 1 Assembled G-quadruplex frame-related DNA sequences
[0031]
[0032]
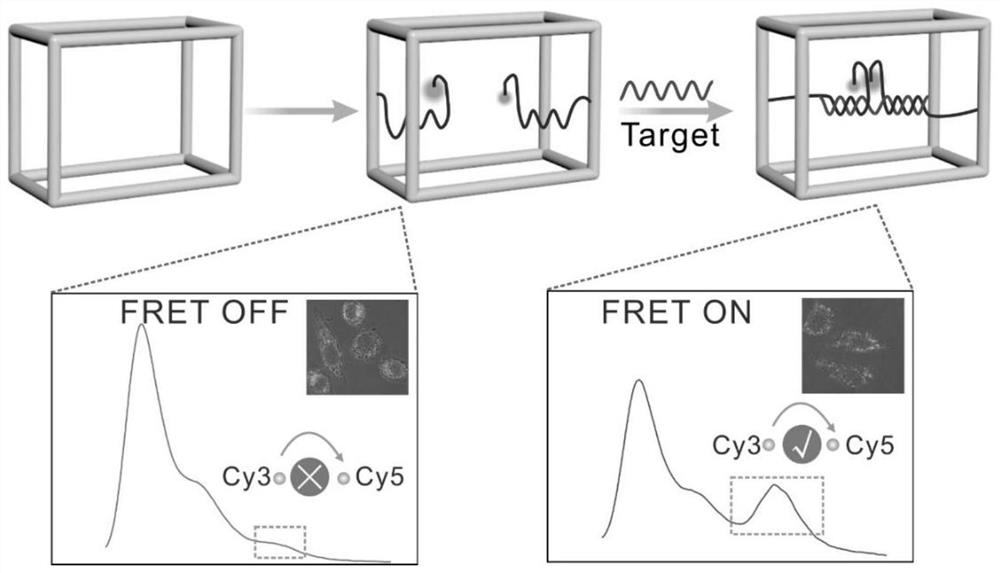
[0033] Then mix evenly in 100 μL of 1×TAMg buffer at a final concentration of 0.6 μM, put the centrifuge tube into a PCR machine and cool down from 95 °C to 4 °C, and the cooling time lasts for 3.5 hours, and the DNA frame-wrapped G- The quadruplex structure, such as figure 1 shown.
Embodiment 2
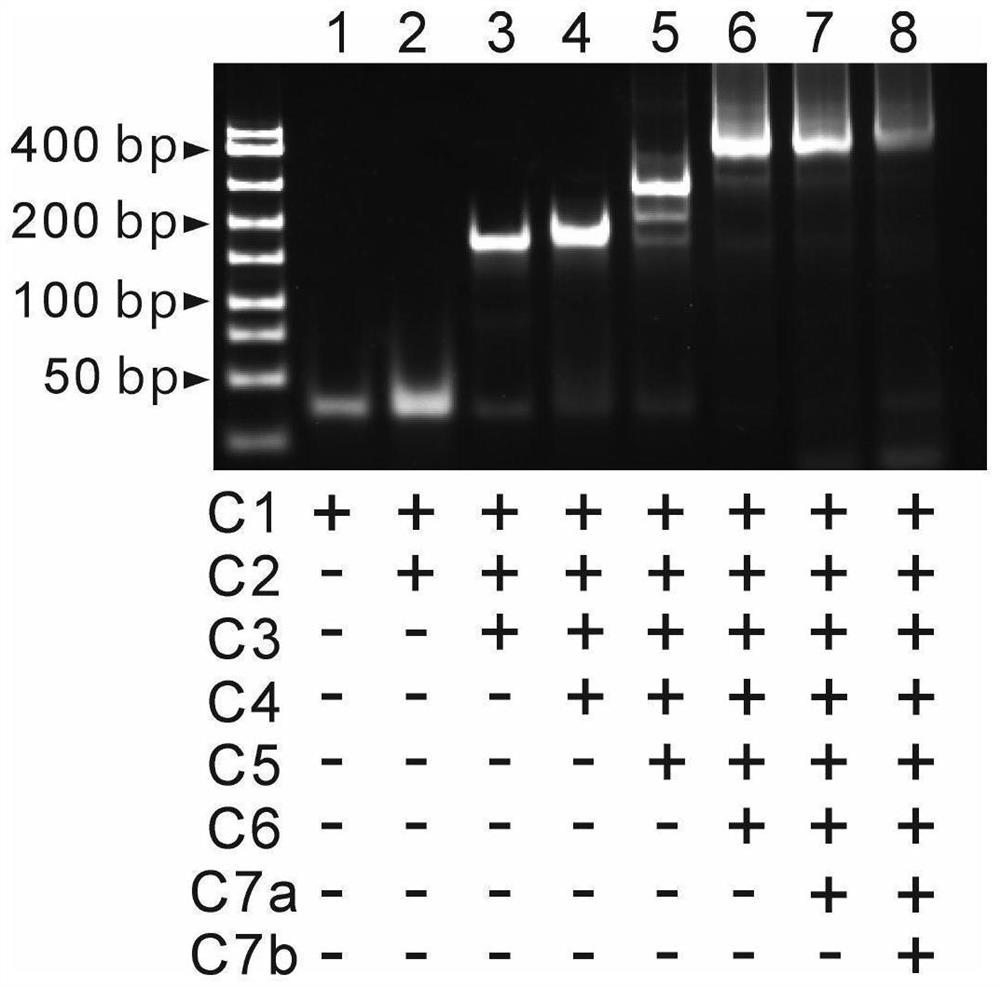
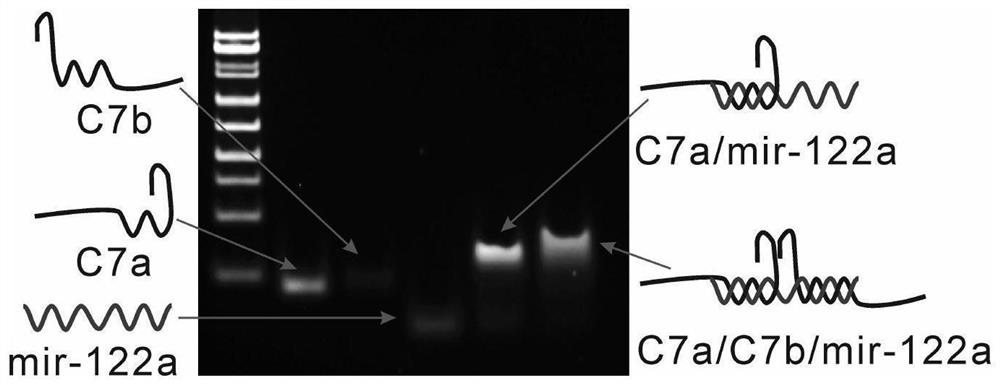
[0035] Prepare 6% PAGE gel with 5×TBE1.2mL, 30% Arc.Bis.1.2mL, three-distilled water 3.6mL, ammonium persulfate 60μL, TEMED6μL, and let it solidify at room temperature for 30 minutes. Eight strands, seven strands, six strands, five strands, four strands, three strands, two strands and single strands forming the tetrahedral DNA nanoframe were annealed to obtain samples with corresponding structures. Add 2 μL of loading buffer to every 10 μL of sample solution, mix well and add 10 uL to the sample well, run in 1xTBE buffer at 90V for 1 hour, then stain the gel in 1% gel-red aqueous solution for 10 minutes, Finally developed in the imager. from figure 2 As seen in , as the number of bands increased, the mobility gradually decreased, indicating the correct assembly of G4-F. In order to verify the ability of the target to hybridize to the split G-quadruplex, the sequences corresponding to C7a, C7b, miR-122a, C7a / miR-122a and C7a / C7b / miR-122a were annealed to obtain samples of th...
Embodiment 3
[0037] After the mica was incubated with 40 μL ((3-aminopropyl)triethoxysilane, APTES) (0.5%) for 5 min, the mica was rinsed with Milli-Q water and rinsed with N 2dry. Incubate 10 μL of samples dissolved in buffer on the treated mica for 5 min. After rinsing, add buffer to a total volume of 100 μL. Scan the G4-F structure for AFM imaging in DimensionIcon (Bruker) mode, the results are as follows Figure 4 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com