SNP molecular marker for identifying scutellaria baicalensis produced in Inner Mongolia as well as method and application of SNP molecular marker
A molecular marker, Scutellaria baicalensis technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial determination/inspection, etc., can solve the problems of low technology scalability and low accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Sample information: Inner Mongolia (HQ361, HQ362, HQ401, HQ402, HQ431, HQ432), Liaoning-Jilin (HQ481, HQ482, HQ501, HQ502, HQ541, HQ542), Shaanxi-Shanxi-Shandong (HQ351, HQ381, HQ382, HQ391) , HQ392, HQ471, HQ472, HQ491, HQ492) area of Scutellaria baicalensis. The specific information of the samples can be found in Table 2.
[0063] Identification steps:
[0064] 1. Sample DNA extraction
[0065] The mCTAB method (modified CTAB method) was used to extract DNA from the leaves of Scutellaria baicalensis in each sample. The specific extraction steps are as follows:
[0066] (1) Put 100 mg of fresh plant leaves into a 2.0 mL centrifuge tube, add a small amount of quartz sand and a magnetic bead at the same time, immerse the fresh leaves in liquid nitrogen for about 1 min, and grind them in a grinder until they are fine. powder ready for use;
[0067] (2) Add 1.5 mL buffer A (0.1 M Tris-HCl pH 8.0; 5 mMEDTA; 0.25 M NaCl; 1 % PVP-40) solution to the ground material, an...
Embodiment 2
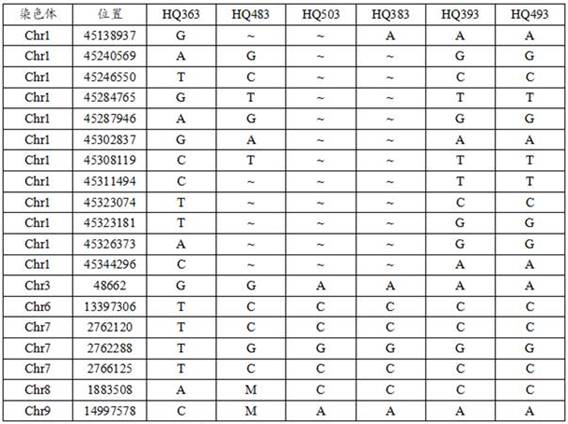
[0091] Sample information: Scutellaria baicalensis were collected from Inner Mongolia (HQ363), Liaoning-Jilin (HQ483, HQ503), Shaanxi-Shanxi-Shandong (HQ383, HQ393, HQ493). The specific information of the samples can be found in Table 2.
[0092] Table 2 Specific information of samples
[0093]
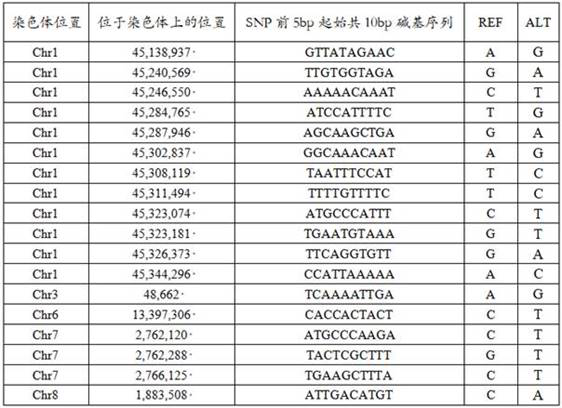
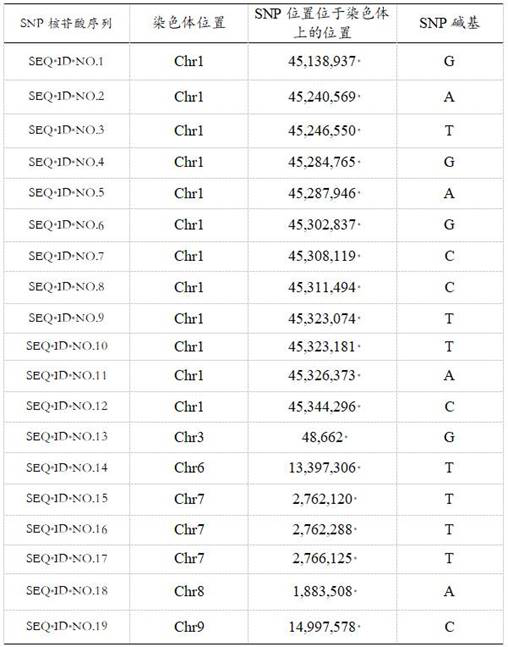
[0094] The SNP sites in Table 1 in Example 1 were used to identify Scutellaria baicalensis in Inner Mongolia area and Scutellaria baicalensis in other regions. Refer to the SNP site information acquisition method in Example 1 to obtain the SNP site information of each sample at the same position.
[0095] The SNP genes at the same positions in each sample were compared with the SNP genes of Scutellaria baicalensis in Inner Mongolia in Table 1, and the comparison results are shown in Table 3.
[0096] Table 3 SNP gene identification results
[0097]
[0098] Note: M stands for CA heterozygote. A single A or C or G or T represents homozygosity.
[0099] From the identification...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com