Method for rapidly detecting RNA viruses based on CCP-FRET
A nucleic acid and sample technology, applied in the field of rapid detection of RNA viruses, can solve the problems of restricting large-scale application, high detection cost, complicated preparation process, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
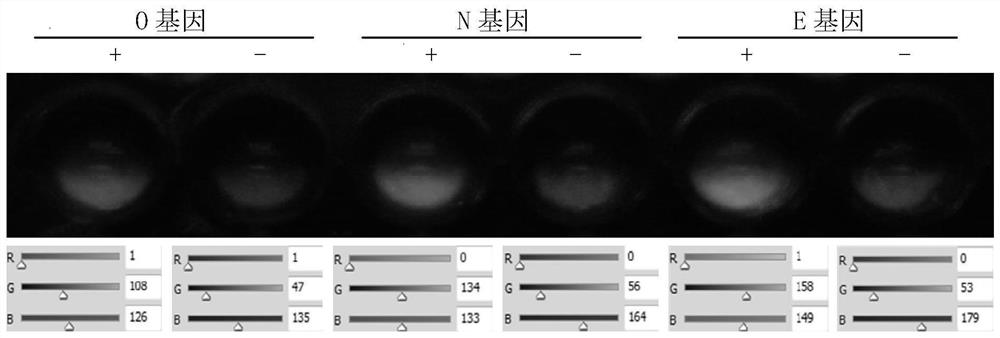
[0102] Example 1. Design and preparation of primers for detecting COVID-2019
[0103] The primers were preliminarily designed through a large number of sequence analysis, and the primers with the best performance were obtained through pre-experimental screening.
[0104] The primer pair for detecting the ORF1ab region gene (O gene) of COVID-2019 is as follows (amplification length 119bp):
[0105] Upstream primer (sequence 1): 5'-CCCTGTGGGTTTTACACTTAA-3';
[0106] Downstream primer (SEQ ID NO: 2): 5'-ACGATTGTGCATCAGCTGA-3'.
[0107] The primer pair for detecting the nucleocapsid protein gene (N gene) of COVID-2019 is as follows (amplification length 99bp):
[0108] Upstream primer (SEQ ID NO: 3): 5'-GGGGAACTTCTCCTGCTAGAAT-3';
[0109] Downstream primer (SEQ ID NO: 4): 5'-CAGACATTTTGCTCTCAAGCTG-3'.
[0110] The primer pair for detecting the envelope protein gene (E gene) of COVID-2019 is as follows (amplification length 113bp):
[0111] Upstream primer (SEQ ID NO: 5): 5-AC...
Embodiment 2
[0114] Embodiment 2, establishment of method
[0115] 1. Prepare a reaction solution in a 200 μl PCR tube (see Table 1 for the components and their addition amounts, and the total volume is 25 μl) for reaction.
[0116] Table 1
[0117] components Amount added For sample solution 2μl dNTP Mixture 2μl upstream primer solution 1μl Downstream Primer Solution 1μl 5×Prime Script II Buffer 4μl PrimeScript II RTase 1μl HotMaster Taq DNA polymerase 0.5μl 10×HotMaster Taq Buffer 2.5μl RNase Free dH 2 o
Make up to 25μl
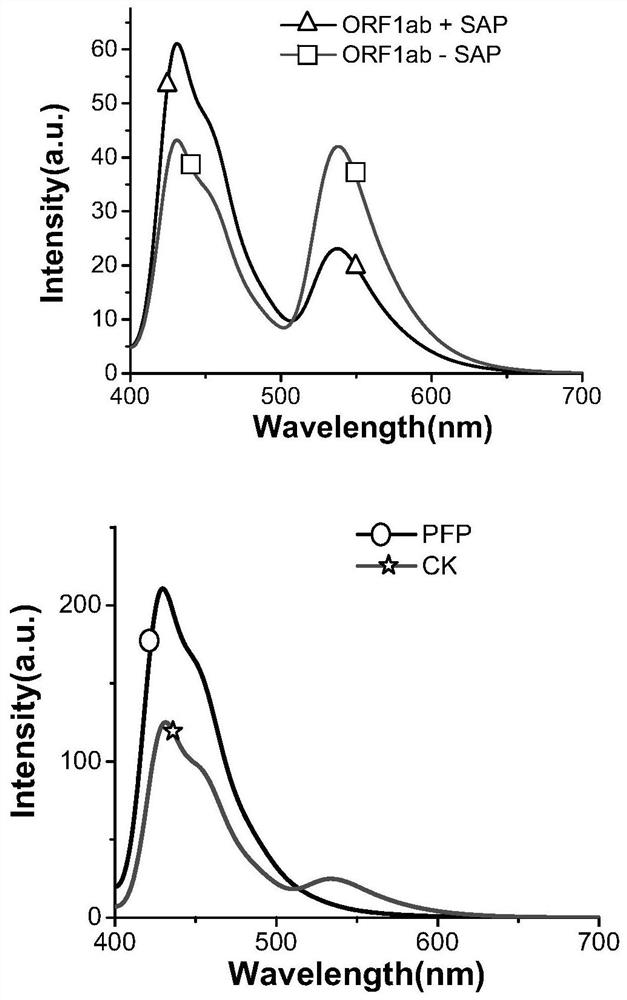
[0118] Active ingredients contained in dNTPs mixture and their concentrations: 100 μM dATP, 100 μM dCTP, 87.5 μM dGTP, 87.5 μM dTTP, 12.5 μM Fl-dGTP and 12.5 μM Fl-dUTP. Fl-dGTP is fluorescein-labeled dGTP (PerkinElmer, NEL429001EA, Fluorescein-12-dGTP). Fl-dUTP is fluorescein-labeled dUTP (PerkinElmer, NEL413001EA, Fluorescein-12-dUTP).
[0119] The upstream primer solution provides a...
Embodiment 3
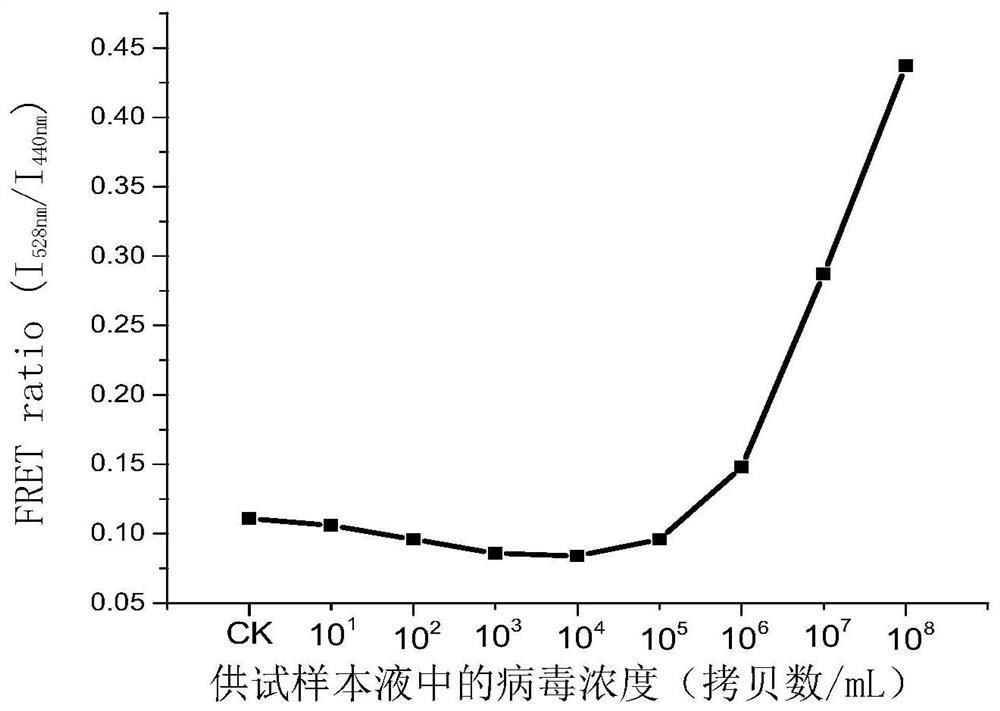
[0132] Sample solution for the test: take armored RNA1 and dilute it with PBS buffer so that the concentration of armored RNA1 is 10 1 Copy number / mL, 10 2 Copy number / mL, 10 3 Copy number / mL, 10 4 Copy number / mL, 10 5 Copy number / mL, 10 6 Copy number / mL, 10 7 copy number / mL or 10 8 Copy number / mL. Control (CK): Use sterile distilled water instead of the sample solution for the test.
[0133] Adopt the method of embodiment 2 to detect. During detection, the primer pair prepared in Example 1 for detecting the O gene of COVID-2019 was used. For detection, use step 3 (1) to detect fluorescence resonance energy transfer.
[0134] see results figure 1 . The minimum virus concentration in the sample solution that can be detected is 10 6 Copy number / mL.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com