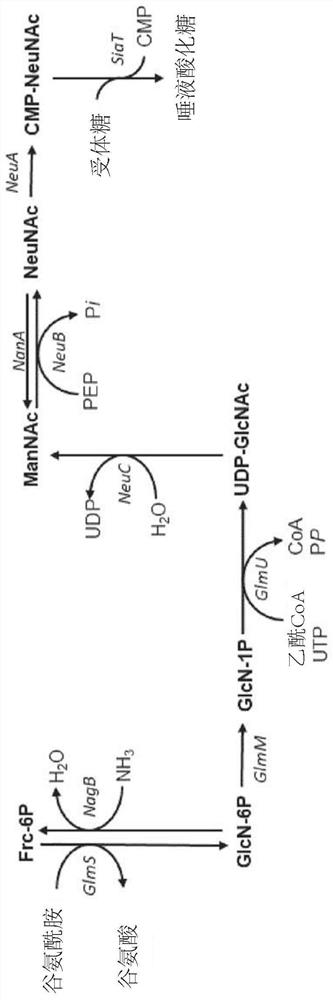
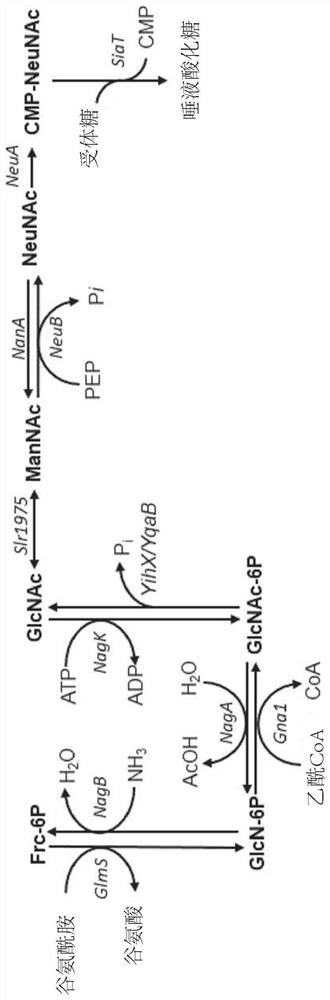
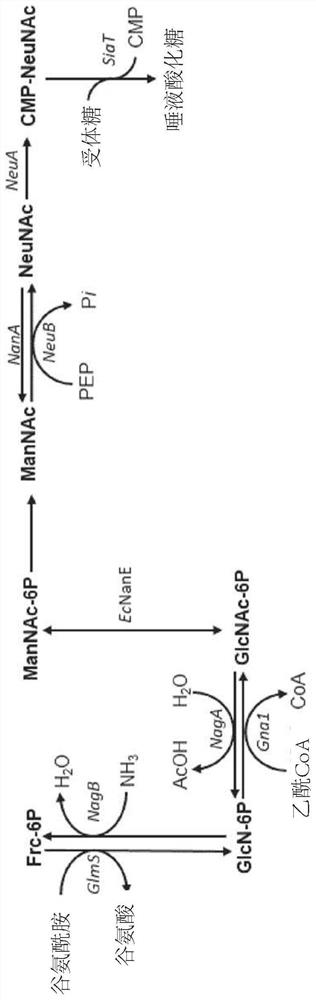
Fermentative production of sialylated saccharides
A technology of sialic acid and sialyltransferase, which is applied in the field of fermentative production of sialylated sugars and compound synthesis, and can solve problems such as deficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0236] Embodiment 1: Production of various sialylated oligosaccharides
[0237] Gene sequences of characterized or putative sialyltransferases were received from literature and public databases. Since sialyltransferases are generally described as showing higher activity when their signal peptide is missing, the inventors used the online prediction tool SignalP (Petersen et al., NatureMethods, 2011Sep 29; 8(10): 785-6) The corresponding protein sequences were analyzed. Genes were synthesized by GenScript Inc. either as annotated full-length forms or, when signal peptides were predicted, as truncated variants lacking the N-terminal signal peptide.
[0238] Sialyltransferases 1 to 26, respectively, were subcloned as operons together with neuA into pDEST14 by SLIC using gene-specific primers, generating a universal type of plasmid: pDEST14-siaT-neuA. The remaining sialyltransferases 27 to 100 were directly subcloned into plasmid pET11a by GenScript using restriction sites NdeI a...
Embodiment 2
[0244] Example 2: Metabolic engineering of Escherichia coli BL21 (DE3) strain for the production of N-acetylneuraminic acid
[0245] Metabolic engineering is achieved through mutagenesis and deletion of specific endogenous genes, as well as genomic integration of heterologous genes. The genes lacZ and araA were inactivated by mutagenesis using mismatch oligonucleotides as described by Ellis et al. (Proc. Natl. Acad. Sci. USA 98:6742-6746 (2001)).
[0246] Genomic deletions were generated according to the method of Datsenko and Wanner (Proc. Natl. Acad. Sci. USA 97:6640-6645 (2000)). To prevent the degradation of N-acetylglucosamine, the following genes were deleted from the genome of E. coli strain BL21(DE3): N-acetylglucosamine-specific PTS enzyme II (nagE), N-acetylglucosamine-6- Phosphate deacetylase (nagA) and glucosamine-6-phosphate deaminase (nagB). Encodes N-acetylmannosamine kinase (nanK), N-acetylmannosamine-6-phosphate epimerase (nanE), N-acetylneuraminic acid aldo...
Embodiment 3
[0256] Example 3: Generation and cultivation of microbial cell lines for the production of 3'-sialyllactose
[0257] Genomic integration tet -siaT9-P t5 -neuA-lox66-aacC1-lox71> (SEQ ID NO: 103) further modified the strain E. coli #NANA1 to generate a 3'-SL production strain. The gene siaT9 (Genbank: BAF91160), codon-optimized for expression in E. coli and synthetically prepared by GenScript, encodes an α-2,3-sialyltransferase from Vibrio sp. JT-FAJ-16. The gene neuA (Genbank: AF305571) encodes a CMP-sialic acid synthase from Campylobacter jejuni.
[0258] Cultivation of strains was performed in 96-well plates. Therefore, a single colony of the strain was transferred from the agar plate to a microtiter plate containing 200 μL of minimal medium containing 7 g l -1 NH 4 h 2 PO 4 、7g l -1 K 2 HPO 4 、2gl -1 KOH, 0.3g l -1 Citric acid, 5g l -1 NH4Cl, 1ml l -1 Antifoam, 0.1mM CaCl 2 , 8mM MgSO 4 , trace elements and 2% sucrose (as carbon source). Trace elements co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com