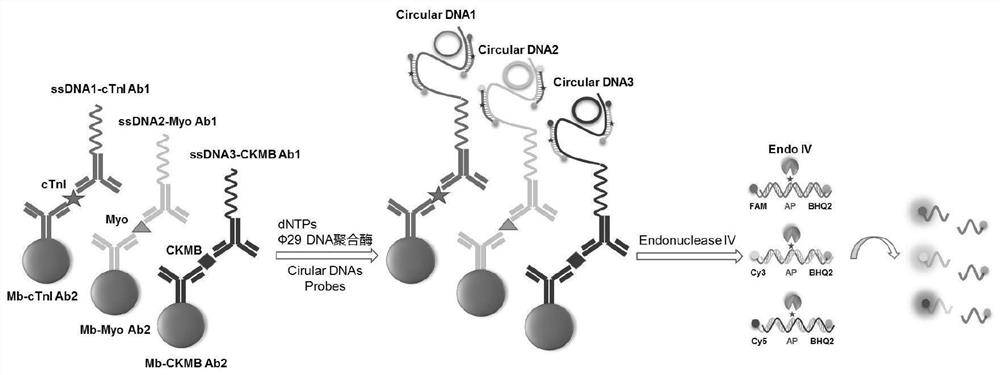
Kit for simultaneously detecting multiple biomarkers based on nucleic acid rolling circle amplification reaction
A rolling circle amplification reaction and biomarker technology, applied in the field of marker detection, can solve the problems of inability to jointly detect multiple biomarkers at the same time, long reaction time, cumbersome operation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0034] The preparation methods of ssDNA1-cTnIAb1, ssDNA2-Myo Ab1 and ssDNA3-CKMB Ab1 in the specific antibody solution in component 1 of the present invention are the same, so only the preparation method of ssDNA1-cTnI Ab1 will be described in detail. The preparation method of ssDNA3-CKMB Ab1 will not be repeated here. The preparation method of ssDNA1-cTnIAb1 of the present invention preferably includes:
[0035] (1) Synthesize ssDNA1 with DIBO group modified at the 5' end to obtain DIBO-ssDNA13;
[0036] (2) carrying out diazotization treatment to cTnIAb1, so that the sugar chain on the antibody Fc fragment is modified to generate a diazo group to obtain N3-cTnIAb1;
[0037] (3) carry out coupling reaction with described DIBO-ssDNA1 and N3-cTnI Ab1, obtain described ssDNA1-cTnIAb1;
[0038] Step (1) and step (2) do not have a time sequence relationship.
[0039] The nucleotide sequence of ssDNA1 of the present invention is preferably shown in SEQ ID NO. 1: AAGTATTACCAGAAAC...
Embodiment 1
[0068] The preparation method of each component in the kit
[0069] 1. Preparation method of component 1:
[0070] 1.1 Preparation of ssDNA1-cTnIAb1
[0071] ①Synthesis of single-stranded DNA1 (DIBO-ssDNA1, SEQ ID NO.1) with modified DIBO group at the 5' end;
[0072] ②Using SiteClick of ThermoFisher TM Antibody Azido Modification Kit treats cTnIAb1, so that the sugar chain on the Fc fragment of the antibody is modified to form a diazo group;
[0073] 3. The single-stranded DNA1 (DIBO-ssDNA1) with the modified DIBO group at the 5' end is reacted and coupled with the diazotized cTnIAb1 (N3-cTnIAb1) to generate ssDNA1-cTnIAb1;
[0074] Table 1 Coupling system and reaction conditions
[0075]
[0076] ④ After the reaction, use molecular sieve to purify to obtain ssDNA1-cTnIAb1.
[0077] 1.2 Labeling method of ssDNA2-Myo Ab1
[0078] ① Synthesize single-stranded DNA2 with DIBO group modified at the 5' end (DIBO-ssDNA2, SEQ ID NO.2);
[0079] ②Using SiteClick of ThermoFi...
Embodiment 2
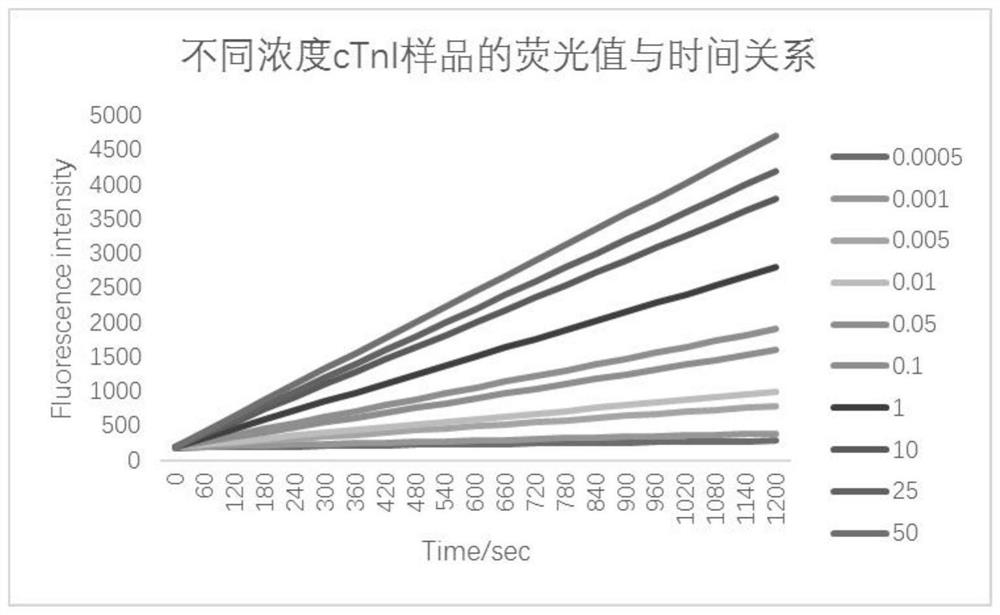
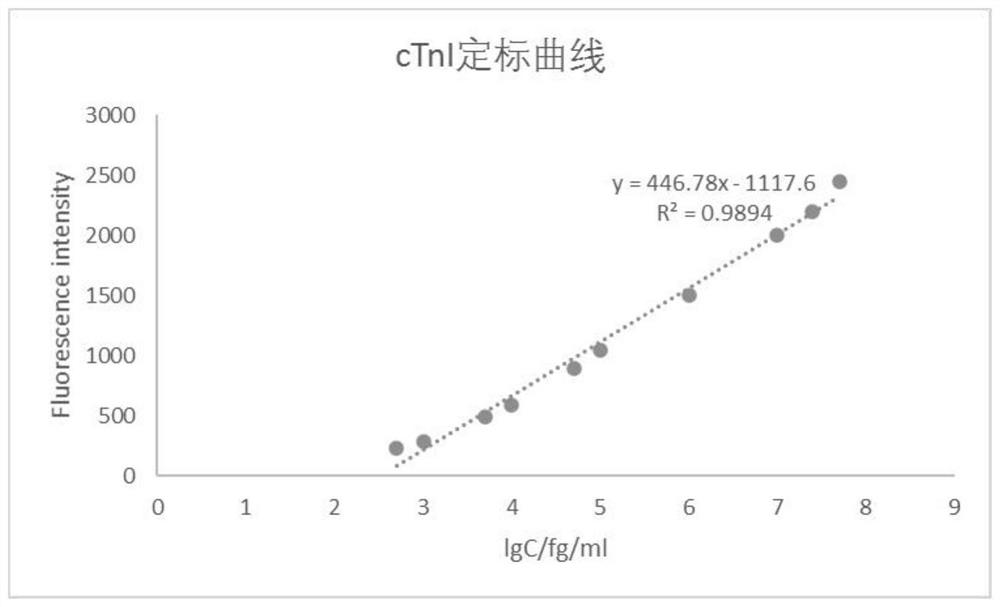
[0141] Sample testing and evaluation
[0142] 1. Sample Preparation
[0143] 1.1 cTnI sample preparation
[0144] The pure cTnI was prepared into samples with different concentration gradients with PBS buffer, the concentrations were 0.0005, 0.001, 0.005, 0.01, 0.05, 0.1, 1, 10, 25, 50ng / ml;
[0145] 1.2 Myo sample preparation
[0146] The pure Myo product was prepared into samples with different concentration gradients with PBS buffer, the concentrations were 0.1, 1, 5, 10, 50, 100, 500, 1000ng / ml;
[0147] 1.3 CKMB sample preparation
[0148] Take pure CKMB and prepare samples with different concentration gradients with PBS buffer, the concentrations are respectively 0.1, 0.5, 1, 2, 5, 10, 20, 50, 100, 200ng / ml;
[0149] 1.4 Preparation of cTnI, Myo, CKMB composite serum samples
[0150] A total of 20 samples from patients with myocardial infarction in the Department of Cardiology and healthy samples for physical examination were taken, and hs-cTnI, Myo, and CKMB were d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com