Method for identifying transcription factor binding element in cotton fiber development period by DNA affinity protein sequencing
A cotton fiber and transcription factor technology, applied in the field of plant molecular biology, can solve the problems of time-consuming and laborious, without considering DNA methylation, aggravating the difficulty of screening functional sites, etc., to shorten the experimental period, strengthen the Effects of practice utilizing value and meaning
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] A method for using DNA affinity protein sequencing to identify transcription factor binding elements during cotton fiber development, specifically adopting the following steps:
[0045] S1. Cotton genomic DNA was randomly interrupted and connected with Illumina sequencing adapters to construct an Index-DNA library
[0046] 1. Random interruption of cotton genomic DNA
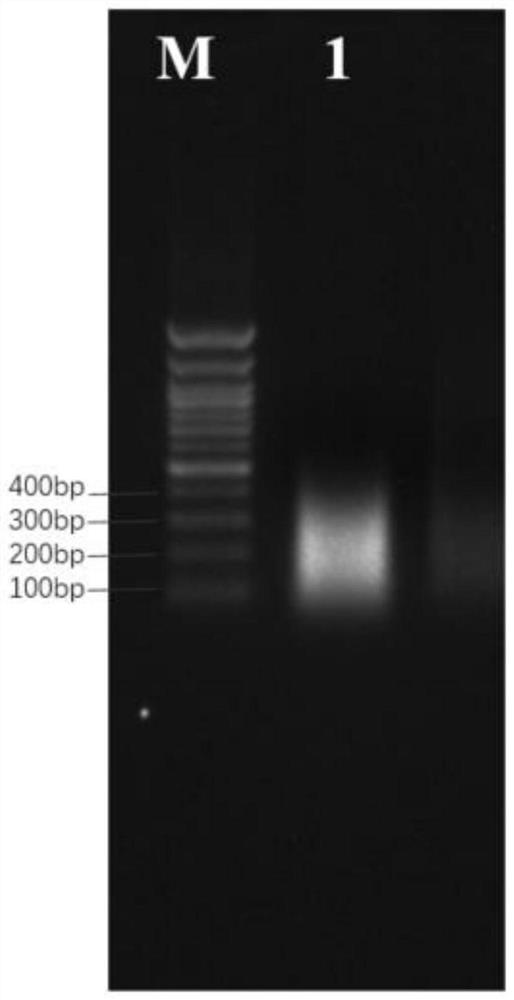
[0047] (1) Use DNeasy Plant Mini Kit (50) plant DNA extraction kit to extract and detect the genomic DNA of cotton fiber development stage. The extraction steps and detection will not be repeated here. Dilute the extracted genomic DNA to 20ng / μL with TE, take 300μl and transfer it to the Covaris microTUBE, and use the ultrasonic breaker of Bioruptor pico to set Time ON 30s Time OFF 30s, Cycle Num 13 Randomly interrupted, after electrophoresis detection, the results are as follows figure 1 As shown, M in the figure represents the DNA marker, and the swimming lanes 1 and 2 are the cotton fiber DNA after i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com