Method for screening phosphoglycerate kinase and substrate binding mode
A combination technology of phosphoglycerate kinase and substrate, which is applied in the fields of bioinformatics, informatics, instruments, etc., to achieve the effect of accurate prediction, screening and improvement of accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
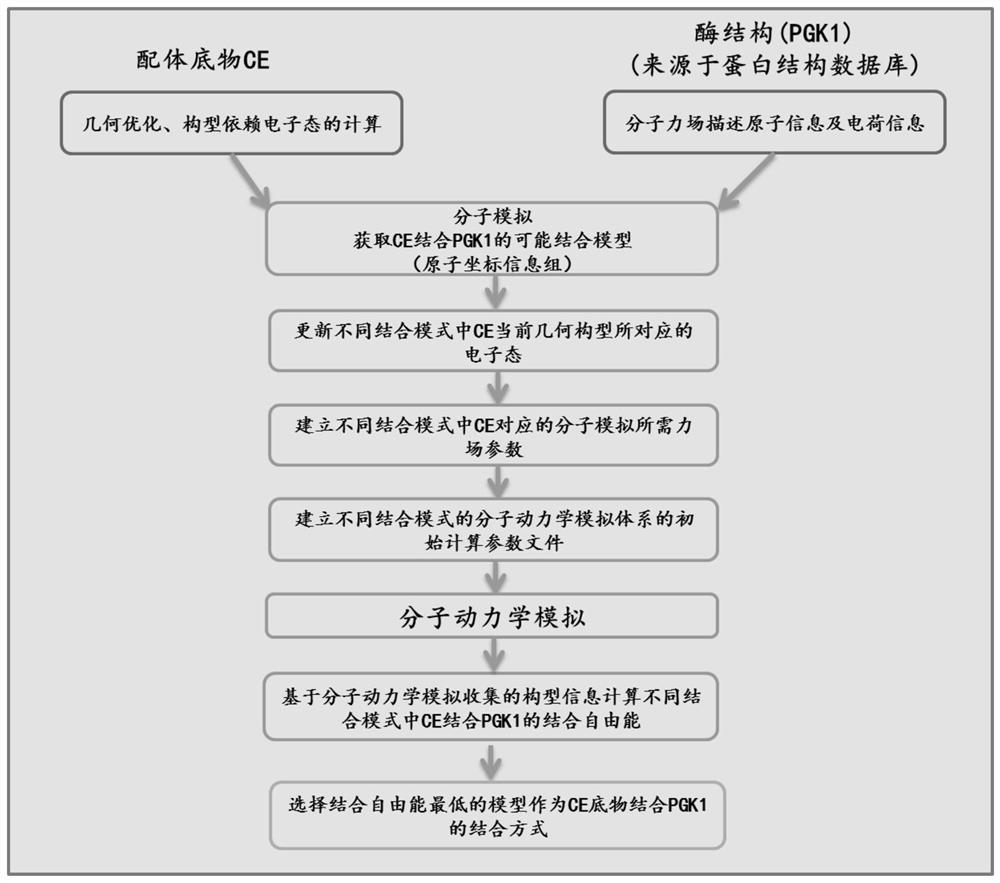
[0034] Example 1 (screening and determination of PGK1 and CE binding mode):
[0035] 1. Prepare the structural information of the predicted substrate CE
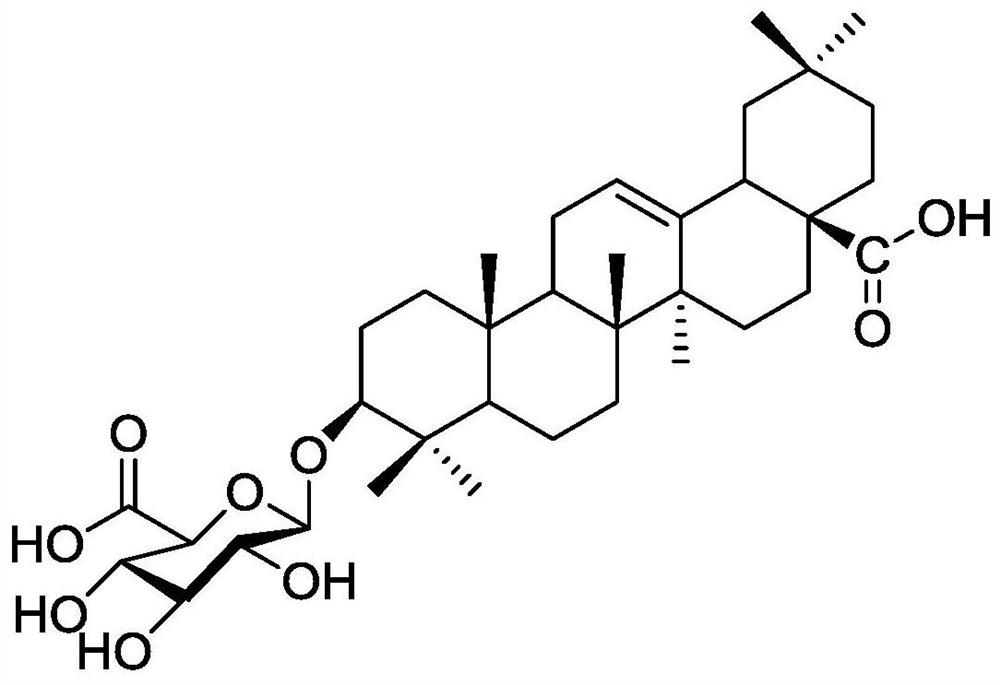
[0036] The structural information of the predicted substrate CE comes from the open source database (https: / / pubchem.ncbi.nlm.nih.gov), query ID: 176079, and its two-dimensional structure is attached figure 2 shown. The atomic types contained in the structure and their corresponding coordinate information were imported into the quantum chemical calculation software gauss 09D01 (M.J.Frisch, et al., Gaussian 09, revision D.1.2010: Gaussian, Inc., Wallingford, CT.), and the density general The B3LYP method is used to optimize the calculation of quantum chemistry under the basis set conditions of 6-31+g* (the basis set parameters come from https: / / www.basissetexchange.org / , query keywords: 6-31+G*), optimize Obtain the calculation result of energy convergence, which contains the new coordinate information of each atom after t...
Embodiment 2
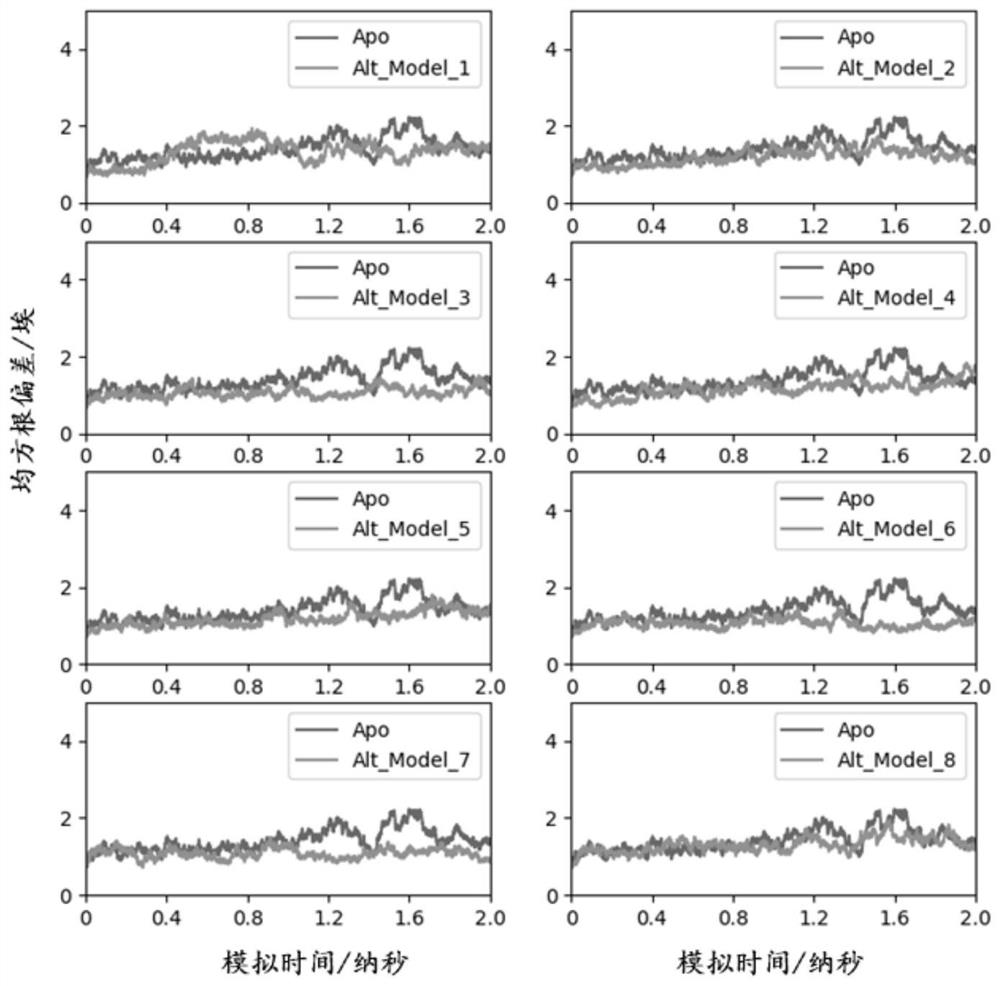
[0266] Example 2 (verification of the binding mode of PGK1 and CE):
[0267] 1. Long-range dynamics simulation of the confirmed binding mode system
[0268]Calculated in Step 7 of Example 1, the No. 7 system determined in Step 9 is calculated through 2ns dynamics simulation, and the new prod_Alt_CE_M7.rst file containing system atom type information and topology information and the original system containing atom type information and atomic coordinates The Alt_CE_M7.top file of information and atomic charge information, the Apo blank control system calculated in step 7, after 2ns dynamics simulation calculation, the new prod_receptor.rst file containing the system atom type information and topology information and the system atom obtained in step 2 The receptor.top file of type information, atomic coordinate information and atomic charge information is used as the input file, and the general dynamics simulation process of constant temperature and constant pressure under the co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com