Method for inhibiting HIV virus replication by targeting reverse transcription primer binding site
A target and inhibitor technology, applied in the field of basic research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
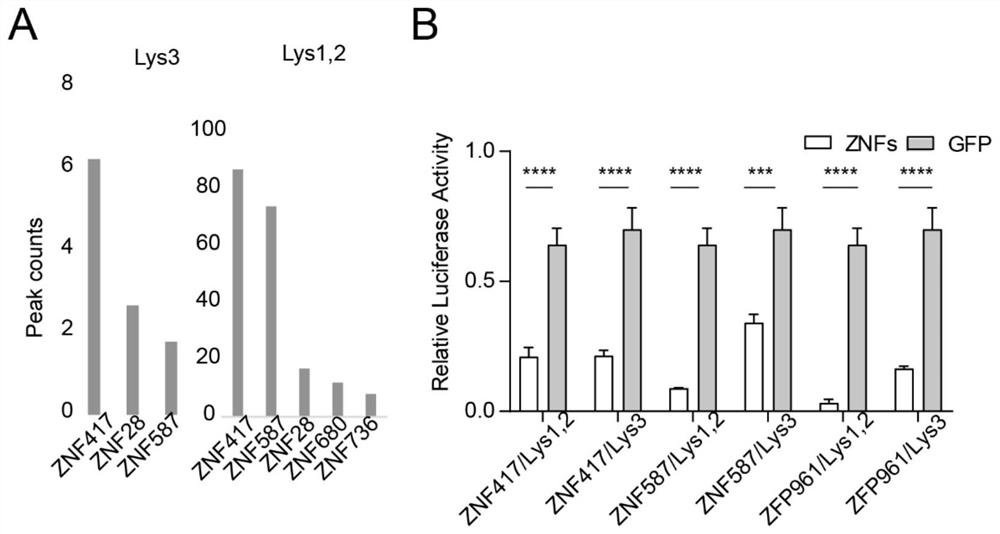
[0049] Example 1: Binding Zinc Finger Proteins The high throughput sequencing database identified ZNF417 and ZNF587 as potential PBS-Lys binding zinc finger proteins.
[0050] 1. Objective: To identify zinc finger proteins that bind HIV target PBS-Lys
[0051] 2. Method:
[0052] 1. By comparing the high-throughput zinc finger protein ChIPseq data of GSE78099 and GSE76496 in the GEO database with the PBS-Lys site, the candidate zinc finger proteins were screened out.
[0053] 2. By inserting the PBS-Lys site before the reporter gene vector promoter, and expressing it together with the zinc finger proteins ZNF417 and ZNF587 expression plasmids, the activity of the reporter gene was detected to verify the binding ability of ZNF417 and ZNF587 to PBS-Lys.
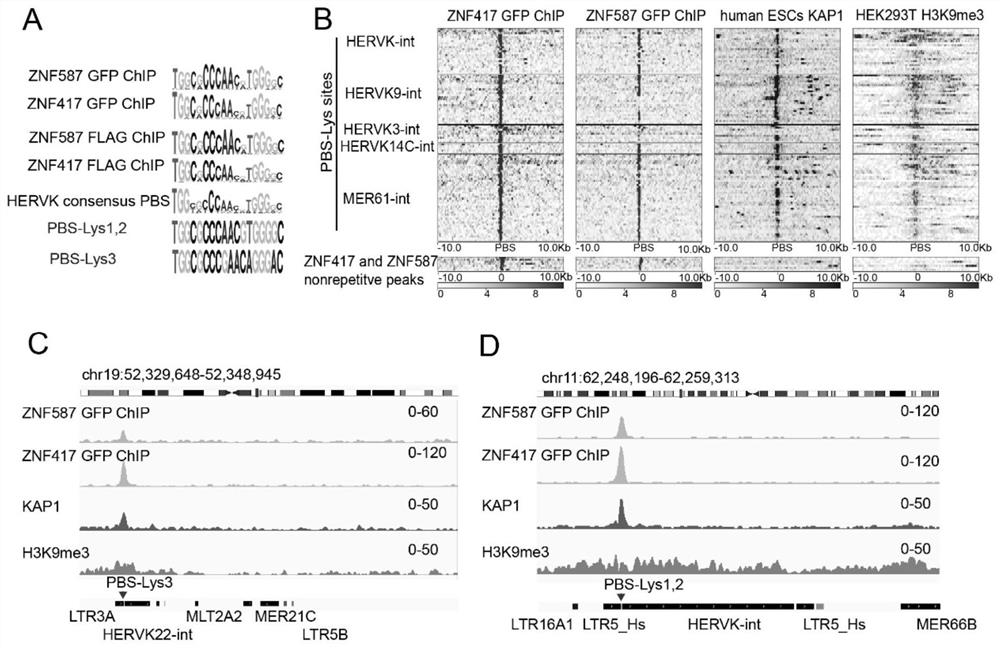
[0054] 3. The inhibitory effect of ZNF417 and ZNF587 on the PBS-Lys site was verified by performing ChIP experiments and high-throughput sequencing after HEK293T cells overexpressed GFP-tag ZNF417 and ZNF587.
[0055] 3. Resu...
Embodiment 2
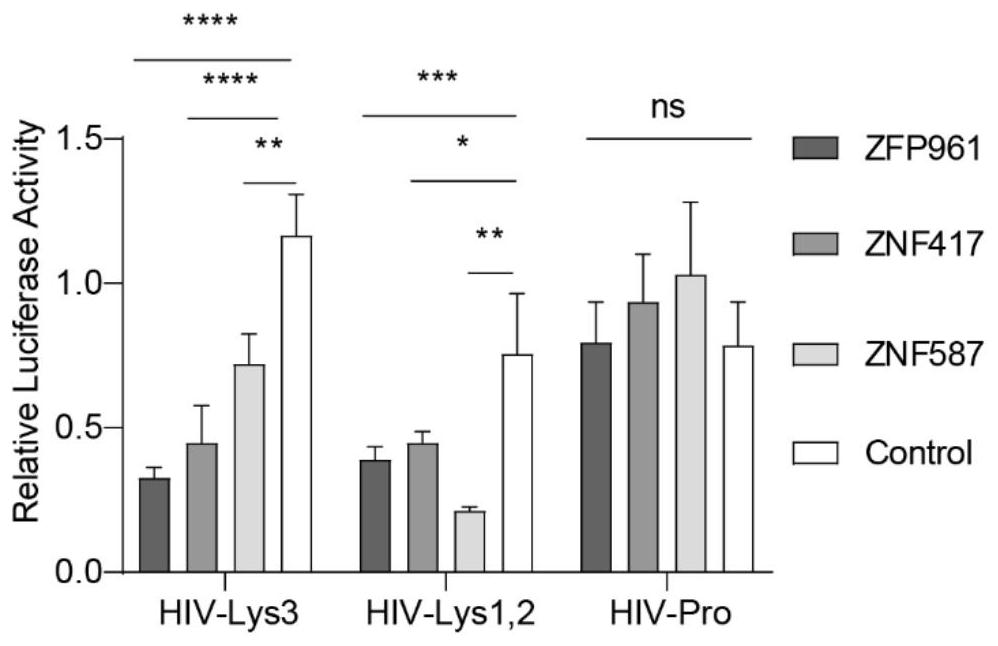
[0057] Example 2: HIV pseudovirus infection experiment to verify the inhibitory effect of ZNF417 and ZNF587 on HIV
[0058] 1. Objective: To verify the ability of ZNF417 and ZNF587 to inhibit HIV
[0059] 2. Method:
[0060] 1. The luciferase reporter gene is driven by the LTR of HIV (similar to the promoter) and co-expressed with ZNF417 and ZNF587 to verify the ability of ZNF417 and ZNF587 to inhibit HIV transcription.
[0061] 2. By designing HIV pseudovirus (here using HIV full-length virus pNL4.3 and backbone virus pSicoR to express RFP, the RFP expression is used to indicate HIV virus infection efficiency) overexpression in ZNF417 and ZNF587 or gene knockout based on CRSIPRcas9 system The infected cells were infected by flow cytometry to determine whether ZNF417 and ZNF587 could inhibit HIV infection.
[0062] 3. By knocking out ZNF417 and ZNF587 in human CD4+ T cells and infecting HIV pseudovirus, simulating the role of ZNF417 and ZNF587 in HIV attacking host cells in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com