Differential screening of deoxyribozyme probe for specifically sensing triple negative breast cancer
A deoxyribozyme probe, triple-negative breast cancer technology, applied in biochemical equipment and methods, instruments, analysis materials, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
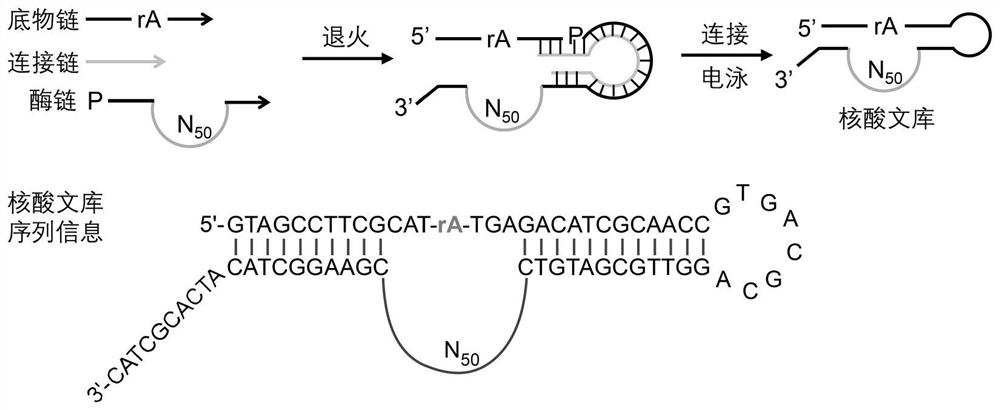
[0035] The construction of embodiment 1 DNA library
[0036] The synthesized substrate chain, enzyme chain and connecting chain were mixed at 1:1:1, incubated at 70°C for 5 minutes, and then annealed at room temperature for 10 minutes; T4 ligase was added and reacted at room temperature for 2 hours. After the reaction, 3 times the volume of -20°C 100% ethanol and 3 μL of 2.5 mg / mL glycogen were added, and placed in a -80°C refrigerator for 10 minutes. The frozen mixture was centrifuged at low temperature and high speed (4°C, 15000rpm, 30min), and the supernatant was removed. After the ethanol volatilized, 20 μL of ultrapure water was added to redissolve, and then subjected to 8% denatured polyacrylamide gel electrophoresis (Denatured Polyacrylamide Gel Electrophoresis, dPAGE ) was purified, and the successfully connected band (full length 119nt) was recovered with a fluorescence imager and a rubber tapping instrument. Add crush-soak buffer (200mM NaCl, 10mM Tris-HCl pH 7.5, 1...
Embodiment 2
[0046] Example 2 Cell culture and extracellular metabolites (extracellular mixtures, EMs) extraction
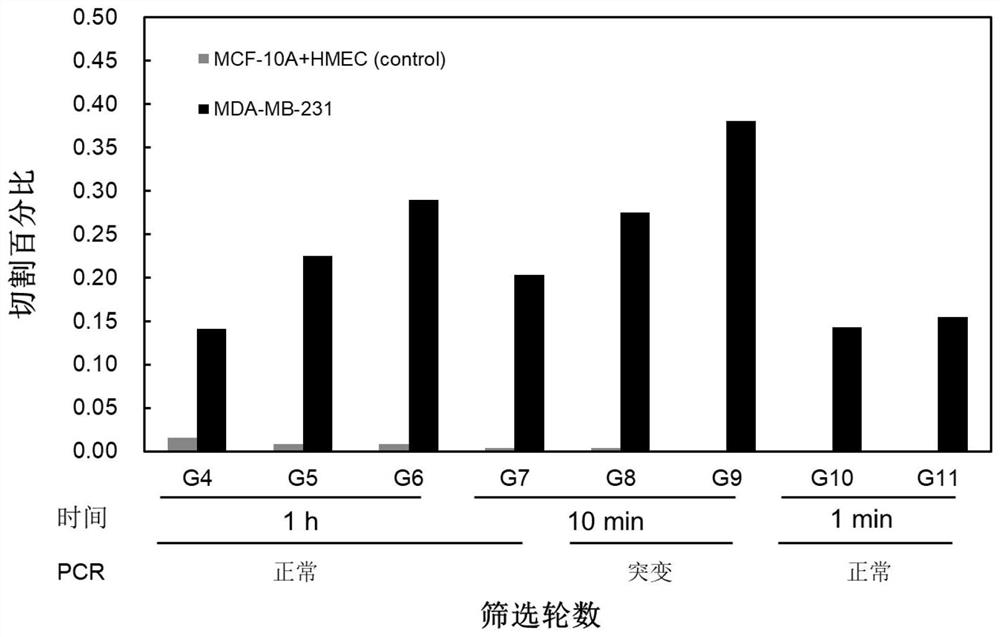
[0047] Select typical TNBC subtype MCA-MB-231 (ATCC No.HTB-26) as the target cell line, normal breast cell line MCF-10A (ATCC No.CRL-10317) and HMEC (ATCC No.PCS-600-010) as control cells. The above-mentioned cells were cultured and passaged according to the culture protocol recommended by ATCC. When the cells were cultured to about 80% confluence, they were washed with PBS three times and then transferred to serum-free medium for 24 hours, centrifuged to remove cell debris and filtered with a 0.22 μm membrane, and protease inhibitors were added to the resulting supernatant, and placed at -80 Store at ℃, and obtain the EMs required for screening.
Embodiment 3
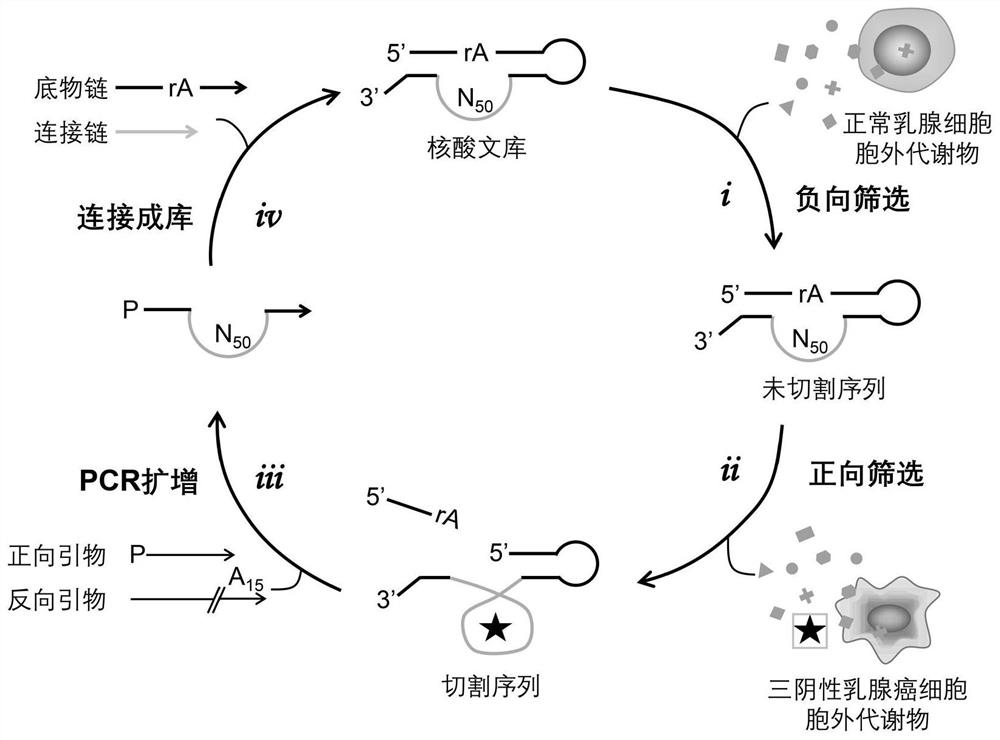
[0048] Example 3 Differential Screening
[0049] Such as figure 2 As shown, the differential screening is divided into steps such as negative screening, positive screening, PCR amplification, and ligation into a library. Clones were sequenced after 11 rounds of selection. Specific screening steps include:
[0050] 1. Negative screening
[0051] Dissolve the DNA library constructed in Example 1 with 75 μL buffer 1, heat in a metal bath at 70°C for 5 minutes, then cool down to 37°C, add 25 μL of EMs of normal mammary epithelial cells MCF-10A and HMEC, 37°C Incubate for 5min; then add 100μL containing Mg 2+ Buffer 2, incubate at room temperature for 1 h for negative selection. Finally, an equal volume of 2×loading buffer (containing urea) was added to terminate the reaction, and the uncut DNA band, ie, the full-length sequence, was separated and purified by 8% dPAGE, and eluted overnight.
[0052]
[0053] 2. Positive screening
[0054] The full-length sequence obtaine...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com