Goat-friendly site SETD5-IN, sgRNA specifically targeting site, and coding DNA and application thereof
A SETD5-IN and friendly site technology, applied in the field of genetic engineering, can solve problems such as system off-target, reduced efficiency of gene-directed editing, and functional impact of off-target sites
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
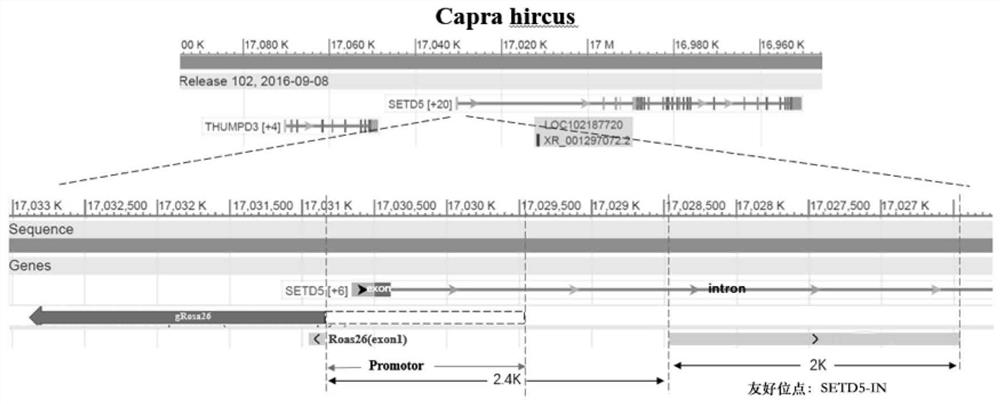
[0077] Example 1. Determination of goat-friendly site SETD5-IN
[0078] The goat-friendly site SETD5-IN is 2kb in length, located on goat chromosome 22, and its position on the goat genome (GCA_001704415.1) is Chr22:17026455-17028455, which is located on the first intron of the SETD5 gene , the sequence is shown as SEQ ID No.1, and its position on the genome is as follows figure 1 shown. The prediction based on the AnimalTFDB database showed that there was no transcription factor binding site in this friendly site; based on the gene position information recorded in the *.gff annotation file of the NCBI existing goat genome, it showed that there was no alternative splicing site in this friendly site. Gene editing within this site will not affect the expression of SETD5 gene and adjacent genes.
Embodiment 2
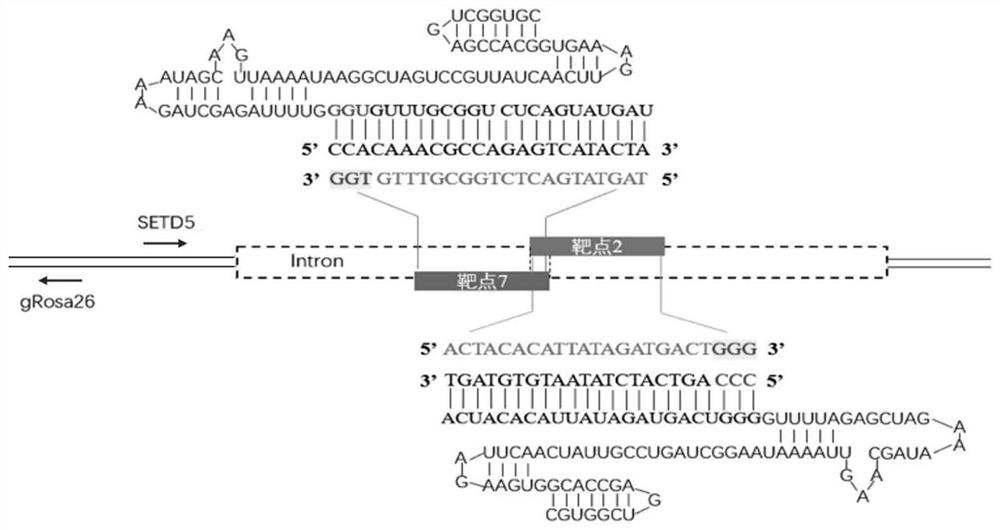
[0079] Example 2, Construction of CRISPR / Cas9 Targeting Vector
[0080] The structure of the CRISPR / Cas9 targeting vector (sgRNA expression vector) is as follows: image 3 shown.
[0081] 1) According to the sgRNA screening principle of the CRISPR / Cas9 system, screen the PAM sequence (5'-NGG) in the goat SETD5-IN site region and the corresponding sgRNA target, and predict the off-target efficiency: select the 13nt at the 5' end of the PAM sequence The sequence was used as the seed sequence and compared with the NCBI online database. Use the sgRNA structure online prediction website (http: / / unafold.rna.albany.edu / q=mfold / RNA-Folding-Form / ) to evaluate the structure and free energy of the sgRNA corresponding to each target, and confirm that the recognition sequence on the sgRNA is not pair with itself.
[0082]The sequence correspondence between the sgRNA target sequence (excluding the PAM region, the sequence in the table below is the positive strand sequence of the genomic ...
Embodiment 3
[0094] Example 3. Verification of gene editing efficiency at the cellular level
[0095] Step 1) Prepare goat fibroblasts. After 40 days of goat pregnancy, the fetus was taken out by cesarean section, and the head, tail, limbs and internal organs were removed, and the tissue was cut into 1mm in size 3 of small pieces. The shredded tissue blocks were evenly placed in the culture dish, and the culture dish was inverted in a cell culture incubator at 38.5°C, 5% CO2, and saturated humidity for 2 hours to make the tissue block adhere to the surface of the culture dish. Add a small amount of DMEM / F12 culture solution containing 10% FBS, continue to culture in the incubator for 24 hours, and add 3 mL of medium to continue the culture. Afterwards, the medium was changed every 3 days, observed every day, and the growth status of the cells was recorded.
[0096] Step 2) The sgRNA expression vector (CRISPR / Cas9 targeting vector) is detoxified and extracted.
[0097] According to the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com