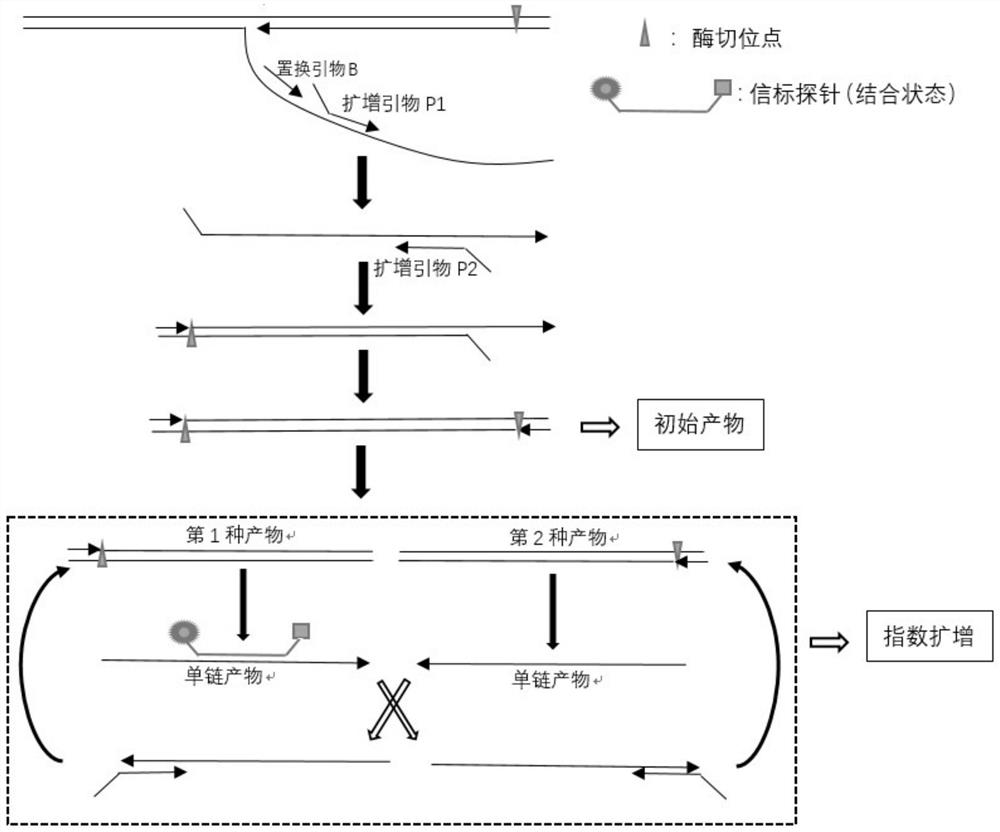
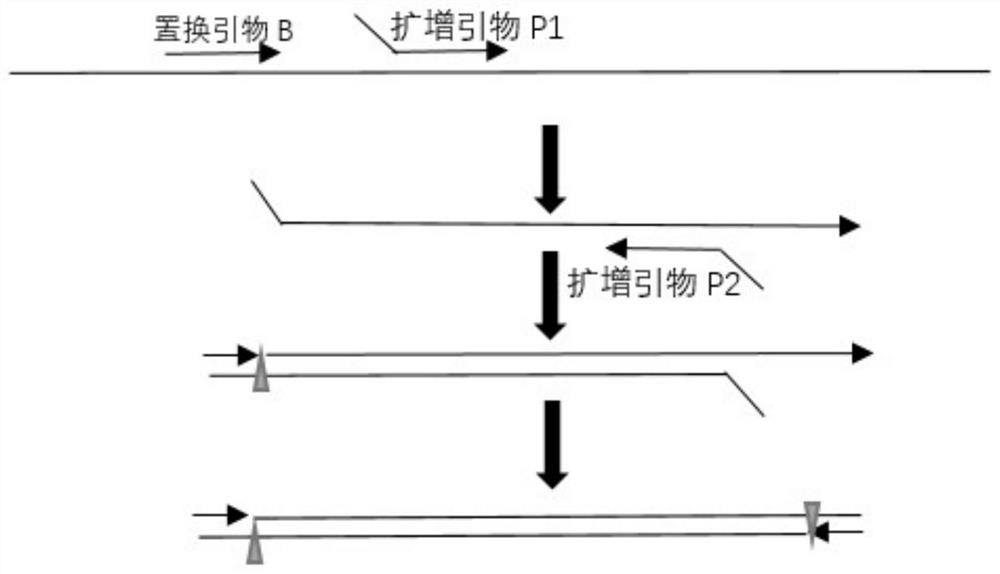
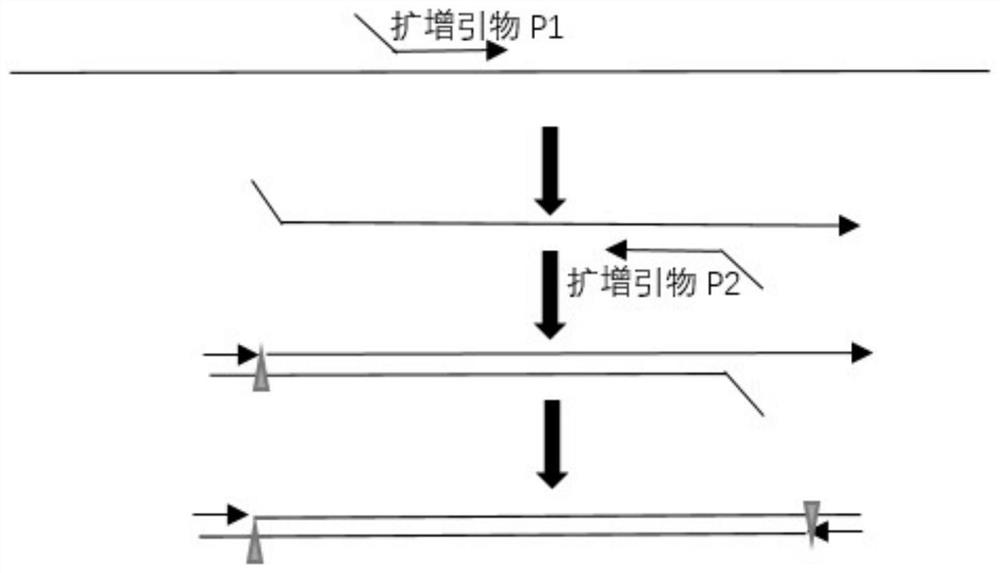
Method for isothermal amplification of nucleic acid target sequence
An isothermal amplification and nucleic acid target technology, which is applied in the field of thermal amplification of nucleic acid target sequences, can solve problems such as false positives, incorrect reaction, unreasonable product analysis, etc., and achieve the effect of ensuring specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0086] The detection of the plasmid that embodiment 1 carries human gene PSMB2
[0087] The primer probe sequence (5'-3') is as follows:
[0088] PSMB2-B (primer): CCCAGCACTTT
[0089] PSMB2-F (primer): TTCAGACTATTGAGTCTATTCTGACCAACAT
[0090] PSMB2-R (primer): GTCAGACTATTGAGTCTTCTCCCAGCTAAT
[0091] PSMB2-P (probe): ATGGTAGTAGAGACGGGGTTTTTACCAT
[0092] The reaction system (final concentration of each component) is as follows:
[0093] Tris-HCl pH8.0, 50mM
[0094] (NH4) 2 SO 4 , 20mM
[0095] MgCl 2 , 10mM
[0096] NaCl, 30mM
[0097] KCl, 10mM
[0098] dNTPs, 1mM
[0099] Betaine, 0.5M
[0100] PSMB2-B, 200mM
[0101] PSMB2-F, 300mM
[0102] PSMB2-R, 300mM
[0103] PSMB2-P, 300mM
[0104] Nt.BstNBI, 3U
[0105] Bst 2.0 warmstart, 4.8U
[0106] The reaction was carried out at 55 ° C, and the signal was collected every 10 s, and the instrument was LightCycler 480II. Detect plasmids 1E5, 1E4, 1E3, 1E2, 1E1 and no template control, the results are as follows...
Embodiment 2
[0107] Example 2 Mycoplasma pneumoniae clinical sample detection
[0108] The primer probe sequence (5'-3') is as follows:
[0109] Mp-B (primer): CTCTCCACTAA
[0110] Mp-F (primer): CATAGACTTATGAGTCTTCTATTCGCTTC
[0111] Mp-R (primer): GTTAGACTTTTGAGTCTTCTTGCTCTGGT
[0112] Mp-P (probe): CGCAGCTGGTTACGGGAATACTGCG
[0113] The reaction system (final concentration of each component) is as follows:
[0114] Tris-HCl pH8.0, 50mM
[0115] (NH4) 2 SO 4 , 20mM
[0116] MgCl 2 , 8mM
[0117] NaCl, 30mM
[0118] KCl, 10mM
[0119] dNTPs, 1mM
[0120] Betaine, 0.5M
[0121] Mp-B (primer), 200mM
[0122] Mp-F (primer), 400mM
[0123] Mp-R (primer), 400mM
[0124] Mp-P (probe), 300mM
[0125] Nt.BstNBI, 3U
[0126] Bst 2.0 warmstart, 4.8U
[0127] The reaction was carried out at 55 ° C, and the signal was collected every 10 s, and the instrument was LightCycler 480II. Detected 8 cases of lung branch samples, the results are as follows Figure 5 As shown, all 8 cases ...
Embodiment 3
[0129] Example 3 Influenza B virus (single-stranded RNA virus) clinical sample detection
[0130] The primer probe sequence (5'-3') is as follows:
[0131] FluB-B (primer): TGTTGCTAAACT
[0132] FluB-F (primer): CTACTGATGAGTCTTTTTAGTGGAGG A T
[0133] FluB-R (primer): CCTTCATTGAGTCTTTTGAAG A GTGA
[0134] FluB-P (probe): ACGGCCATCGGATCCTCAAGCCGT
[0135] Note:" A " Modified with LNA.
[0136] The reaction system (final concentration of each component) is as follows:
[0137] Tris-HCl pH8.0, 50mM
[0138] (NH4) 2 SO 4 , 20mM
[0139] MgCl 2 , 8mM
[0140] NaCl, 30mM
[0141] KCl, 10mM
[0142] dNTPs, 1mM
[0143] Betaine, 0.5M
[0144] FluB-B (primer), 200mM
[0145] FluB-F (primer), 400mM
[0146] FluB-R (primer), 400mM
[0147] FluB-P (probe), 300mM
[0148] Nt.BstNBI, 3U
[0149] Bst 3.0, 6U
[0150] The reaction was carried out at 55 ° C, and the signal was collected every 10 s, and the instrument was LightCycler 480II. Detect 8 clinical samples of i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com