Primers and method for detecting SNP gene typing of AhFAD2B genes of peanuts in high throughput
A genotyping and high-throughput technology, applied in the field of molecular biology, can solve the problem of high detection cost, achieve low detection cost, improve efficiency and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
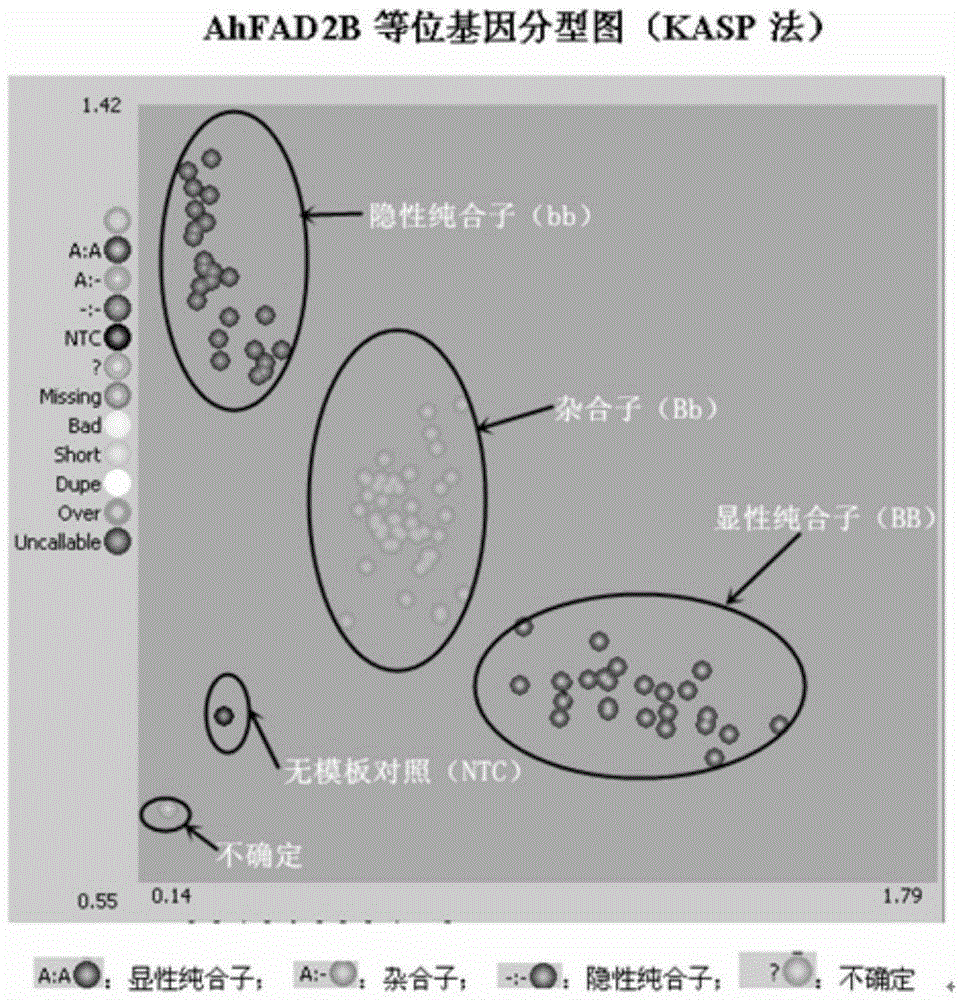
[0033] Example 1. Design of primers for detection of AhFAD2B gene 442 A insertion mutation
[0034] The 442th base sequence of the ORF region of the AhFAD2B gene genome sequence has two sequences: wild type deletion and mutant type insertion of A base, resulting in the mutation of the encoded amino acid sequence from the wild type encoding a complete protein to the mutant type encoding a frameshift and premature termination. According to the position of the above-mentioned mutation site, design KASP allele differential primers (as shown in Table 1, SEQIDNo:1, SEQIDNo:2), so that the differential sequence of the AhFAD2B gene is located at the 3' end of the primer, and the 5' end is connected to the Allele -1tail and Allele-2tail, these two tailing sequences are identical to the sequences of the universal fluorescent probes in the LGC MasterMix detection kit, and the 5' ends of the probes are labeled with FAM and HEX fluorescent groups respectively (the probe sequence Patented b...
Embodiment 2
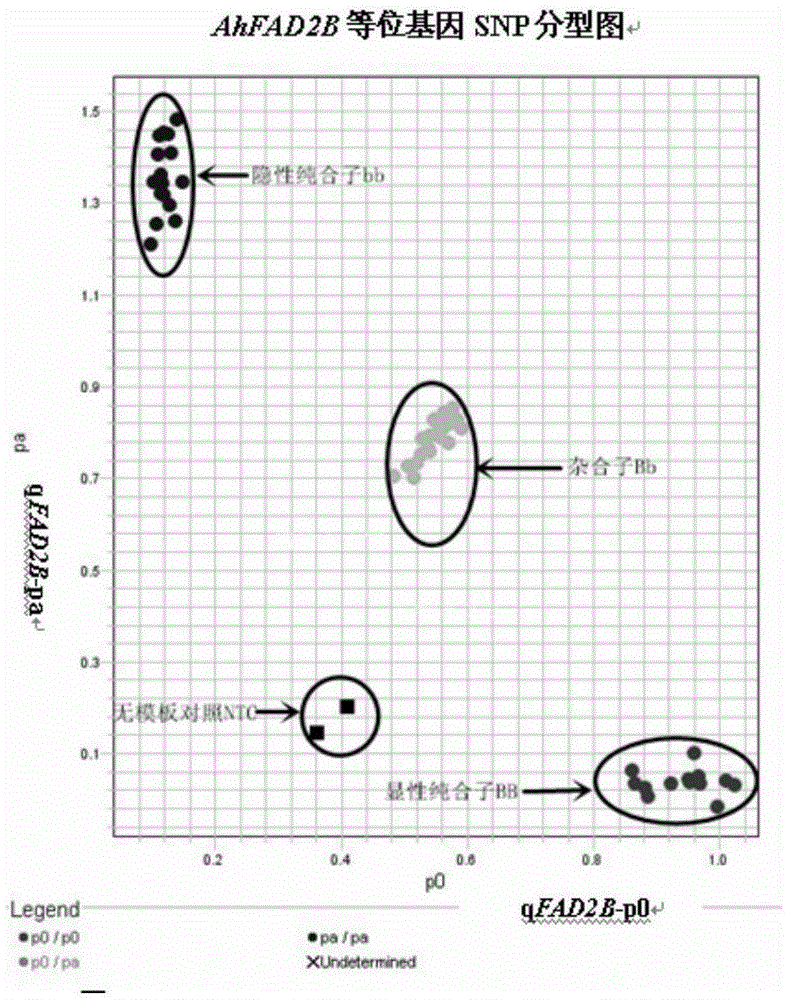
[0040] Embodiment 2, the detection of peanut AhFAD2B gene SNP mutation site
[0041] Allelic site detection was performed with primers designed according to the allelic difference of the AhFAD2B gene and LGC company kit MasterMix probe
[0042] Use the primers and MasterMix probes designed according to the allelic difference of the AhFAD2B gene in Example 2 to test peanut materials, including ordinary oleic acid peanuts (genotype aaBB) as the female parent and high oleic acid peanuts (genotype aabb) as the male parent. The F1 generation hybrids and the F2 generation segregation groups were detected, and the common oleic acid peanuts and high oleic acid peanuts of the female parents were used as controls at the same time. The specific methods are as follows:
[0043] (i) Extraction of genomic DNA from tested peanut samples
[0044] According to the Plant Genomic DNA Extraction Kit (catalogue number: DP-305) provided by TIANGEN Company, and according to the instructions of the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com