Multiple real-time quantitative PCR primer, probe and detection method for identifying viral pathogens relevant to fever with eruption syndrome as infection diseases
A real-time fluorescence quantitative and symptomatic technology, applied in biochemical equipment and methods, microbial determination/inspection, resistance to vector-borne diseases, etc., can solve the problems of untimely detection, insufficient sensitivity, time-consuming and cumbersome, etc., and achieve a high degree of automation , high sensitivity, stable results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1: Design of primers and probes
[0048] By comparing and analyzing the gene sequences of all known viruses, select highly conserved segments, and design multiple pairs of primers and probes. The length of the primers is generally about 20 bases. The optimal primer-probe sequence combination is as follows:
[0049] Analyze the information of the above-mentioned virus-specific genes, and use the sequence analysis software DNASTAR to exclude inter-primer / inner dimers, and verify the specificity of the primers and the homology of similar pathogens by BLAST, and design primers and primers for the detection of the above-mentioned pathogens. probe.
Embodiment 2
[0050] Embodiment 2: Construction and optimization of PCR reaction system
[0051] 1. Extraction of total nucleic acid from the specimen
[0052] Each strain is provided by our laboratory. At the same time, the virus titer titration (TCID 50 / ml), TCID 50 That is, half of the infectious dose of tissue cell culture, each group of virus strains were removed 1 TCID 50 / ml of virus load.
[0053] The inactivated virus was used as the sample to be tested, and the nucleic acid was extracted using the QIAGen RNeasy Mini Kit (Qiagen, Chatsworth, CA, catalog #74104). The operation is carried out according to the instruction manual. Extraction process and main parameters:
[0054] (1) When using this kit for the first time, add 100% ethanol to the AW1 and AW2 buffers according to the volume indicated on the reagent bottle, add 25 ml absolute ethanol to 19 ml AW1, add 30 ml absolute ethanol to 13 ml AW1 Ethanol; add 28 μg / ml carrier RNA to AL.
[0055] (2) Put 25 μl Qiagen Pro...
Embodiment 3
[0077] Example 3: Sensitivity Analysis of Multiplex Fluorescent Quantitative RT-PCR
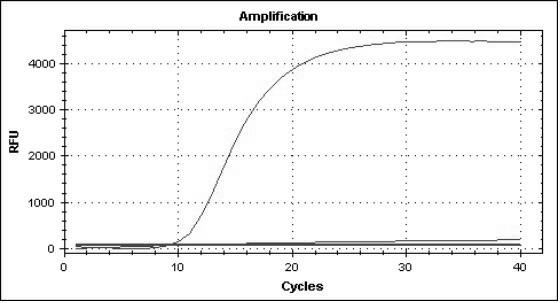
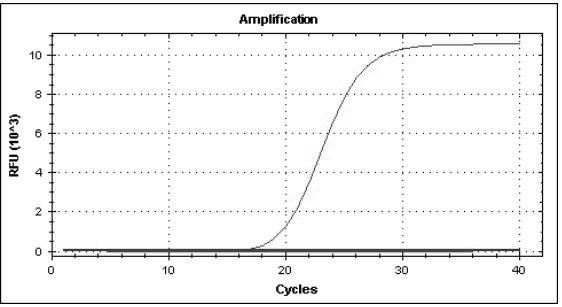
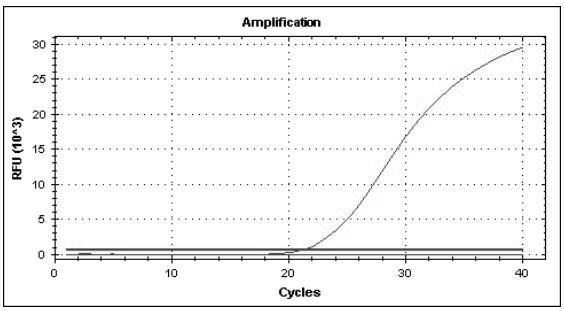
[0078] According to the above method, the virus pathogens of the above fever with rash syndrome infectious diseases (including rubella virus, measles virus, dengue virus, varicella zoster virus, human DNA parvovirus, enterovirus 71, coxsackie virus A16 ) after nucleic acid extraction, use the established multiplex real-time fluorescent quantitative RT-PCR method, and use the same reaction system and cycle parameters for detection and identification. The results showed that each group of reactions had corresponding specific fluorescence amplification curves for the nucleic acid templates of all 8 strains of viruses, and the corresponding templates of other virus strains were all negative ( Figure 1-8 ), which shows that the established multiplex real-time fluorescent quantitative PCR method has good specificity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com