Biomarker and method for DNA sample tracking detection, and application of biomarker
A biomarker, tracking and detection technology, applied in the field of genetic detection and tracking, can solve the problems of inability to cover, increase data, increase the complexity of data processing and detection cost, etc., and achieve the effect of shortening the cycle, accurate results and high cost performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
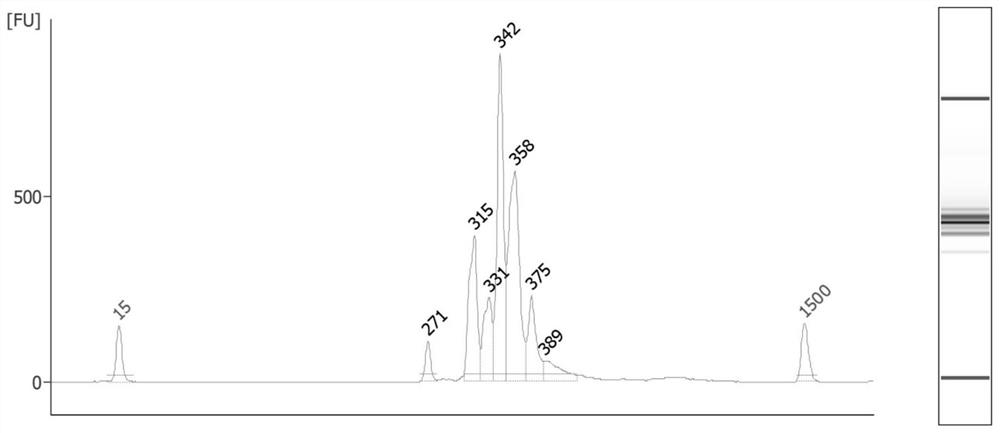
[0044] A total of 16 samples from 4 families and non-family probands were tested using IDT's all-exon panel, A1-A3 was family 1, A4-A6 was family 2, A7-A8 was family 3, and B6-B8 It is family 4, and the remaining samples are independent probands; the default order of adding samples is as shown in the figure below:
[0045]
[0046] The actual sequence of adding samples is shown in the figure below:
[0047]
[0048] The test results found that A1, A2, and A3 do not conform to the family relationship; A4, A5, and A6 do not conform to the family relationship; A7, A8 and B1 do not conform to the family relationship; B6, B7, and B8 do not conform to the family relationship; it is suspected that this batch of samples is mixed up Seriously, use the tracking method of this application to construct a library for this batch of 16 DNA samples in accordance with the preset loading order. Proceed as follows:
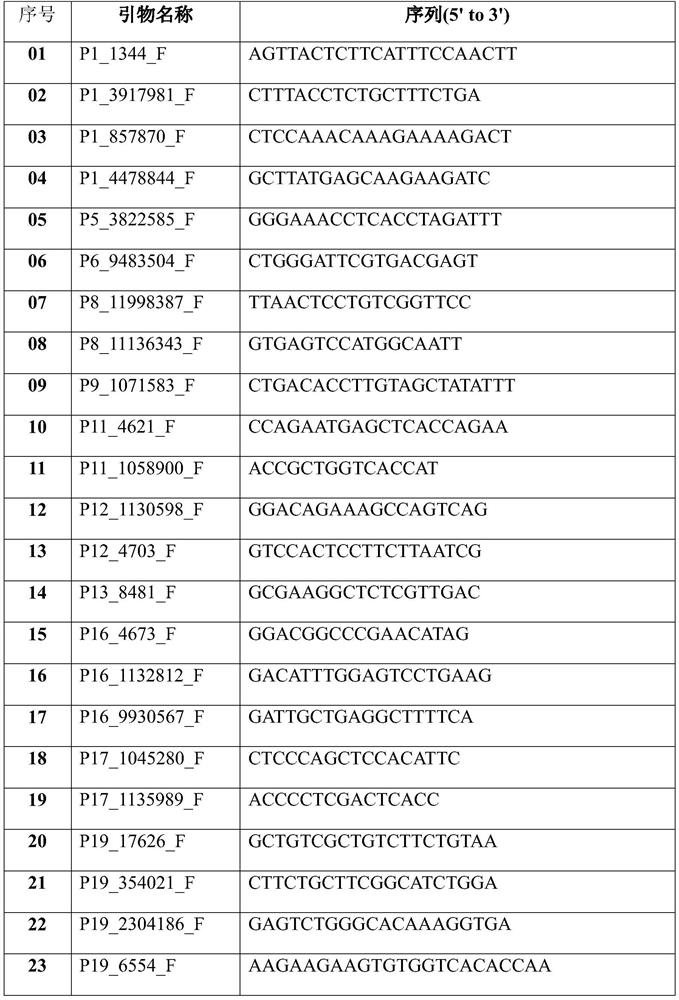
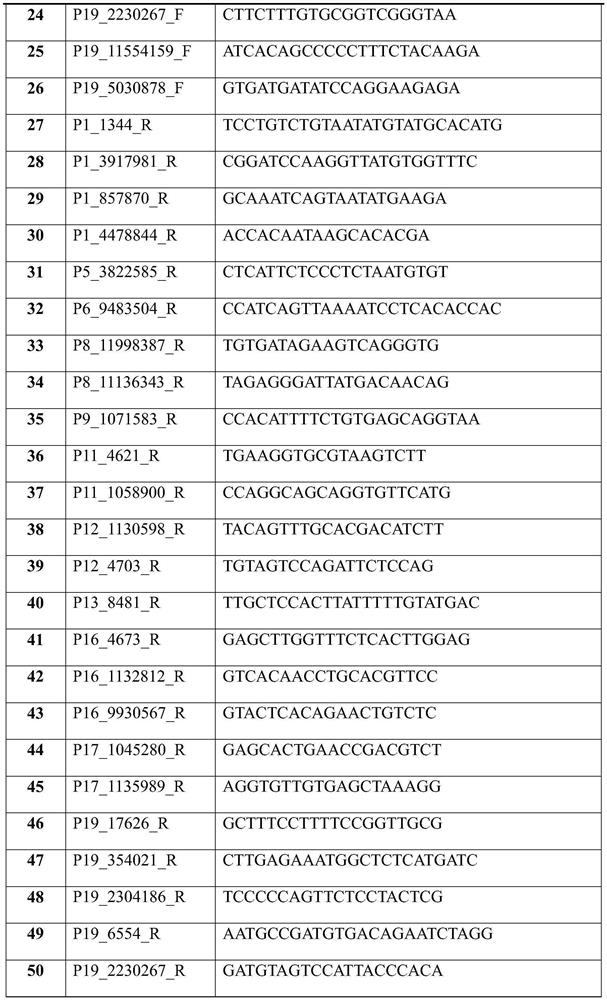
[0049] 1. Multiplex primer mix
[0050] Add 52 single primer master so...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com