T cell antigen receptor, polymer complex thereof, and preparation method and application of polymer complex
A cell antigen and complex technology, applied in the direction of antibody mimics/scaffolds, animal cells, antibody medical components, etc., can solve the problem of insignificant killing effect of T cell receptors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0169] Example 1: Construction and Effect Detection of CMV Antigen Epitope Tetramer
[0170] 1. Construction of CMV epitope tetramer
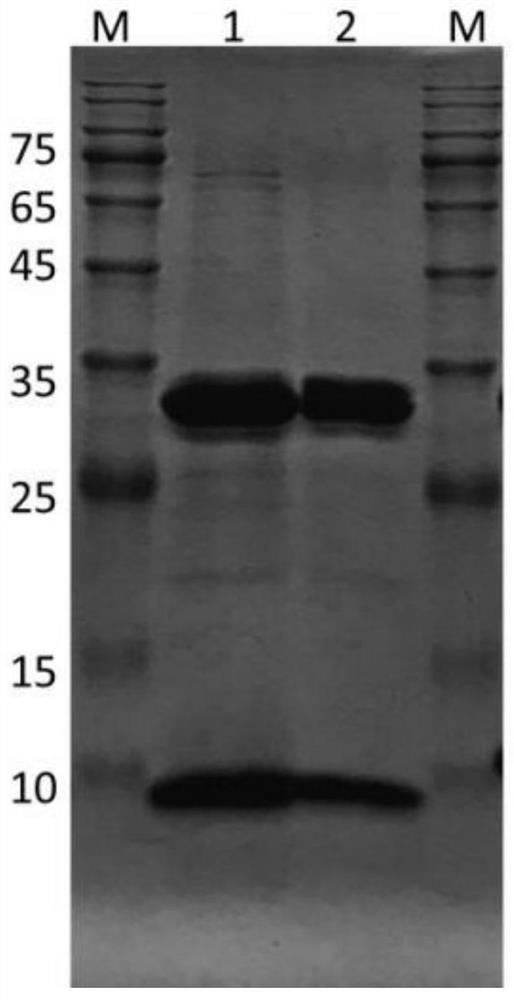
[0171] 1) α chain and β2m chain (whose amino acid sequence is shown in SEQ ID NO: 25) of HLA-A*1101 (whose amino acid sequence is shown in SEQ ID NO: 24 and its nucleotide sequence is shown in SEQ ID NO: 25) expressing sequence optimization NO: 26, the nucleotide sequence is shown in SEQ ID NO: 27). Wherein, the structure of the α-chain is that the extracellular region sequence of the corresponding HLA-type α-chain is linked with the Avi-tag sequence, separated by BamHI restriction sites to provide biotinylation sites. The β2m chain has the signal peptide sequence removed and two amino acids (M and A) added in front of the mature peptide sequence. The expression vector is PET28a+, and the expression strain is transetta or BL21. The concentration of IPTG was 0.5mM, and the expression was induced for 4h. Extraction of α-chain and β2m-catenin ...
Embodiment 2
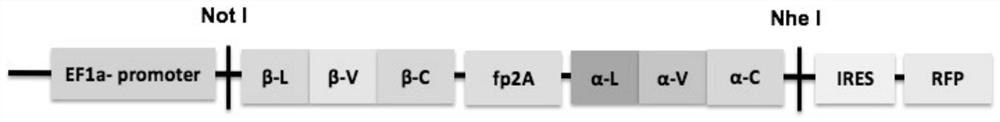
[0187] Embodiment 2: Construction of pHAGE-TCR-RFP carrier
[0188] 1. Obtain β and α gene fragments of CMV pp65 epitope-specific TCR
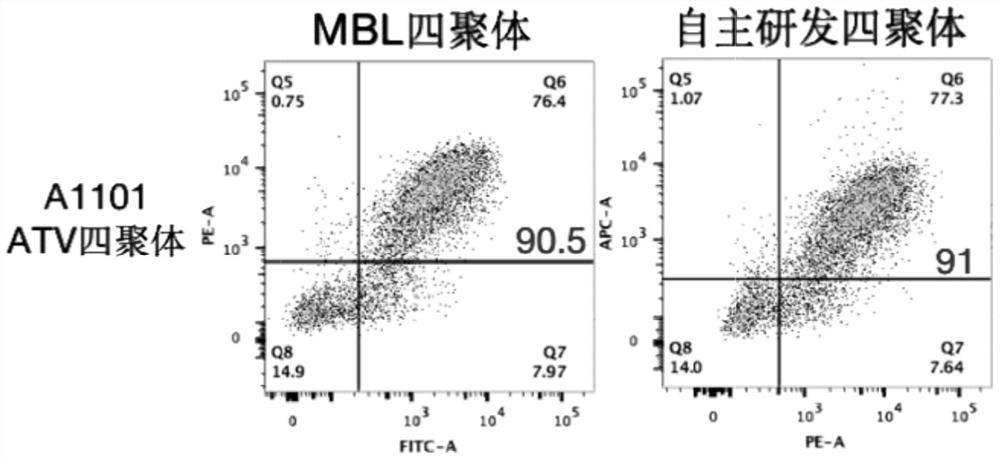
[0189] 1) Use the HLA-A*1101-ATVQGQNLK-tetramer purchased from MBL or made by our company, and stain the peripheral blood according to the product manual, and perform flow cytometry single cell sorting on the T cells positive for the tetramer staining, cDNA was obtained by reverse transcription ( IV Reverse Transcriptase, Invitrogen). According to the multiplex PCR (Multiplex PCR) principle, the variable region fragment of the TCRβ gene was amplified by two rounds of PCR (KOD-Plus-Neo, TOYOBO).
[0190] The reverse transcription primer is: TRBC1-TCAGGCAGTATCTGGAGTCATTG (SEQ ID NO: 136)
[0191] PCR amplification primers are:
[0192] Upstream primer 1: T cell receptor β variable region--TRBV_F1 (SEQ ID NO: 56 to 95, wherein SEQ ID NO: 60, 59 can also be separately used as the variable region fragment of the β gene of C44 and C45 TRBV_F1)...
Embodiment 3
[0211] Example 3: Detection of membrane expression and affinity of TCR by pMHC tetramer staining
[0212] 1. Construction of endogenous TCR knockout JurkatT cell line
[0213] Based on the sequence characteristics of TCR in Jurkat cells, guide sequences were designed in the constant regions of the α chain and β chain (TRA_oligo1-CACCGTCTCTCAGCTGGTACACGGC (SEQ ID NO: 20), TRA_oligo2-AAACGCCGTGTACCAGCTGAGAGAC (SEQ ID NO: 21), TRB_oligo1-CACCGGGCTCCAAACACAGCGACCTC (SEQ ID NO :22), TRB_oligo2-AAACGAGGTCGCTGTGTTTGAGCCC (SEQ ID NO: 23)).
[0214] The guide sequences of the synthesized α-chain and β-chain were respectively constructed into sgRNA-LentiCRISPR-puro and sgRNA-LentiCRISPR-BSD lentiviral vectors, and co-transfected with packaging plasmids psPAX2, pMD2.G and PEI transfection reagents in a certain ratio to 293T Cells, the cell culture supernatants of 48h and 72h were collected, and the concentrated two viruses simultaneously infected the human JurkatT cell line. 48 hours a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com