Primer group, kit and application of primer group in rapid identification of pepper root rot bacteria
The technology of a primer set and a kit, which is applied in the application field of primer sets and rapid identification of pepper root rot bacteria, can solve the problems of inaccurate identification results and complicated operation methods, shorten the time required for identification, simplify the identification process, Improve the effect of anti-efficacy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] A method for rapid identification of prickly ash root rot bacteria (fusarium solani), comprising the following steps:
[0036] S1. Synthetic primer set: the primer set includes upstream primer F1 and downstream primer R1; the sequence of upstream primer F1 is: 5'-CGTGTCGCAGAAGAGGG-3', and the sequence of downstream primer R1 is: 5'-AATAGGAAGCCGCTGAGC-3'; The synthetic concentrations of primer F1 and downstream primer R1 were both 10 μmol / L;
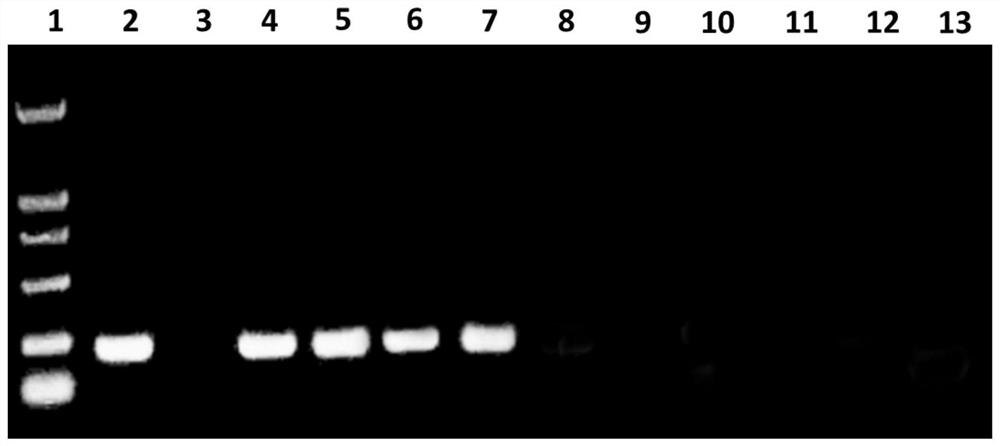
[0037] S2. Extract sample DNA: Using conventional DNA extraction means (CTAB method), extract the concentration of 50ng / μL from Fusarium solani, Fusarium oxysporum, Fusarium graminearum, Fusarium moniliforme and Botrytis cinerea respectively. DNA diluent to obtain multiple sets of sample DNA;
[0038] S3. Prepare the PCR reaction system: add the sample DNA of each group into their respective 25 μL reaction systems, and each 25 μL reaction system includes the following components:
[0039]
[0040]
[0041] S4. Routine PCR d...
Embodiment 2
[0046] A method for rapid identification of prickly ash root rot bacteria (fusarium solani), comprising the following steps:
[0047] S1. Synthetic primer set: the primer set includes upstream primer F1 and downstream primer R1; the sequence of upstream primer F1 is: 5'-CGTGTCGCAGAAGAGGG-3', and the sequence of downstream primer R1 is: 5'-AATAGGAAGCCGCTGAGC-3'; The synthetic concentrations of primer F1 and downstream primer R1 were both 10 μmol / L;
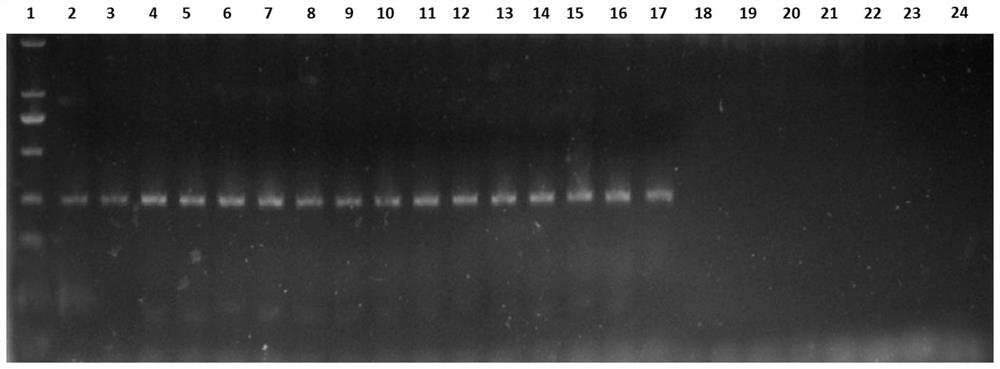
[0048] S2. Extracting sample DNA: using conventional DNA extraction means (CTAB method), extracting a DNA dilution with a concentration of 50 ng / μL from the rhizosphere soil of the pathogenic Zanthoxylum bungeanum root inoculated with Fusarium solani to obtain sample DNA;
[0049] S3. Prepare the PCR reaction system: add the sample DNA into the 25 μL reaction system, which includes the following components:
[0050]
[0051] S4. Routine PCR detection: According to the above-mentioned PCR reaction system, use the sample DNA as a t...
Embodiment 3
[0056] A method for rapid identification of prickly ash root rot bacteria (fusarium solani), comprising the following steps:
[0057] S1. Synthetic primer set: the primer set includes upstream primer F1 and downstream primer R1; the sequence of upstream primer F1 is: 5'-CGTGTCGCAGAAGAGGG-3', and the sequence of downstream primer R1 is: 5'-AATAGGAAGCCGCTGAGC-3'; The synthetic concentrations of primer F1 and downstream primer R1 were both 10 μmol / L;
[0058] S2. Extract sample DNA: Using conventional DNA extraction means (CTAB method), extract the concentration of 50ng / μL from the healthy Zanthoxylum bungeanum roots that have not been inoculated with Fusarium solani and the diseased Zanthoxylum bungeanum roots that have been inoculated with Fusarium solani, respectively. DNA diluent to obtain 2 sets of sample DNA;
[0059] S3. Prepare the PCR reaction system: Add the two groups of sample DNA into their respective 25 μL reaction systems, and the 25 μL reaction systems of each gr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com