DNA methylation sequencing library and construction method and detection method thereof
A technology for constructing methods and sequencing libraries, applied in chemical libraries, biochemical equipment and methods, libraries, etc., can solve the problems of uncorrectable methylation information, data contamination, and distortion of methylation information, so as to avoid redundant sequencing data. extra effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
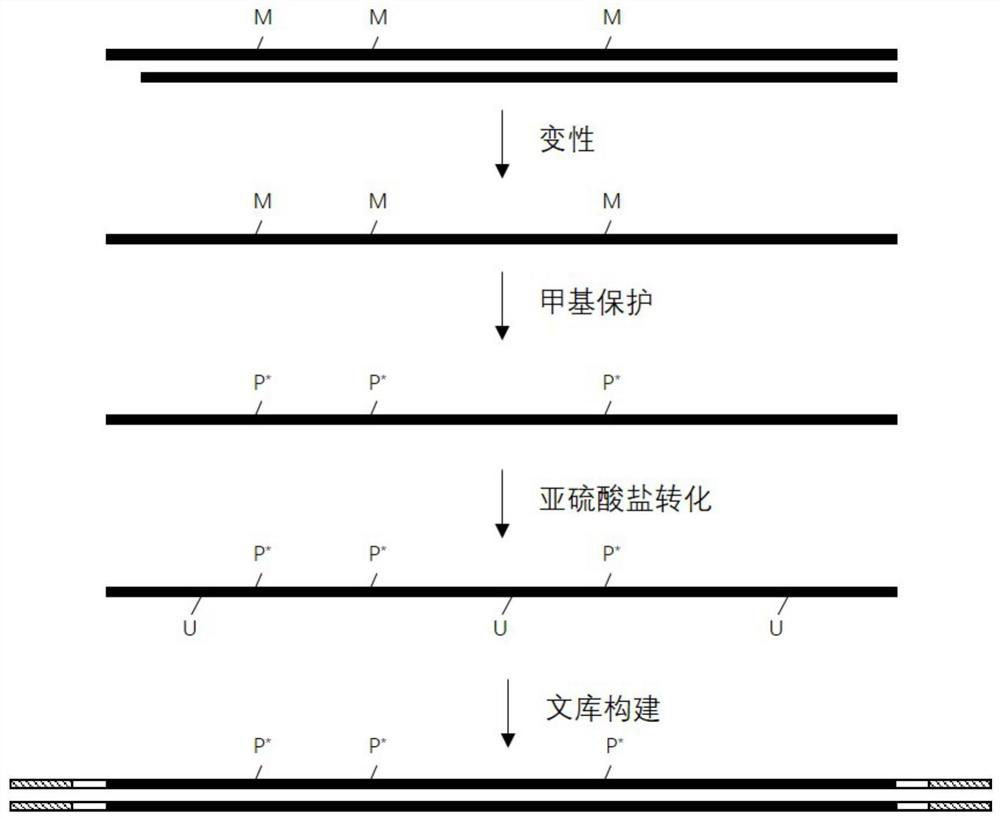
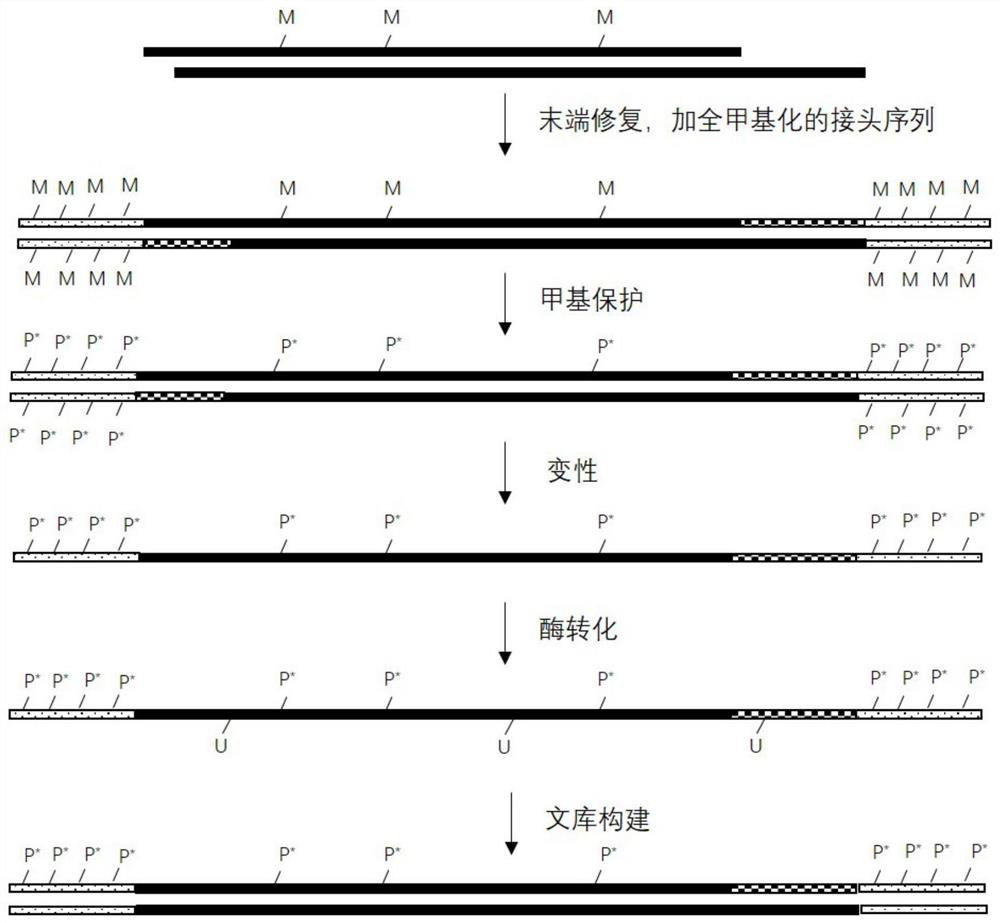
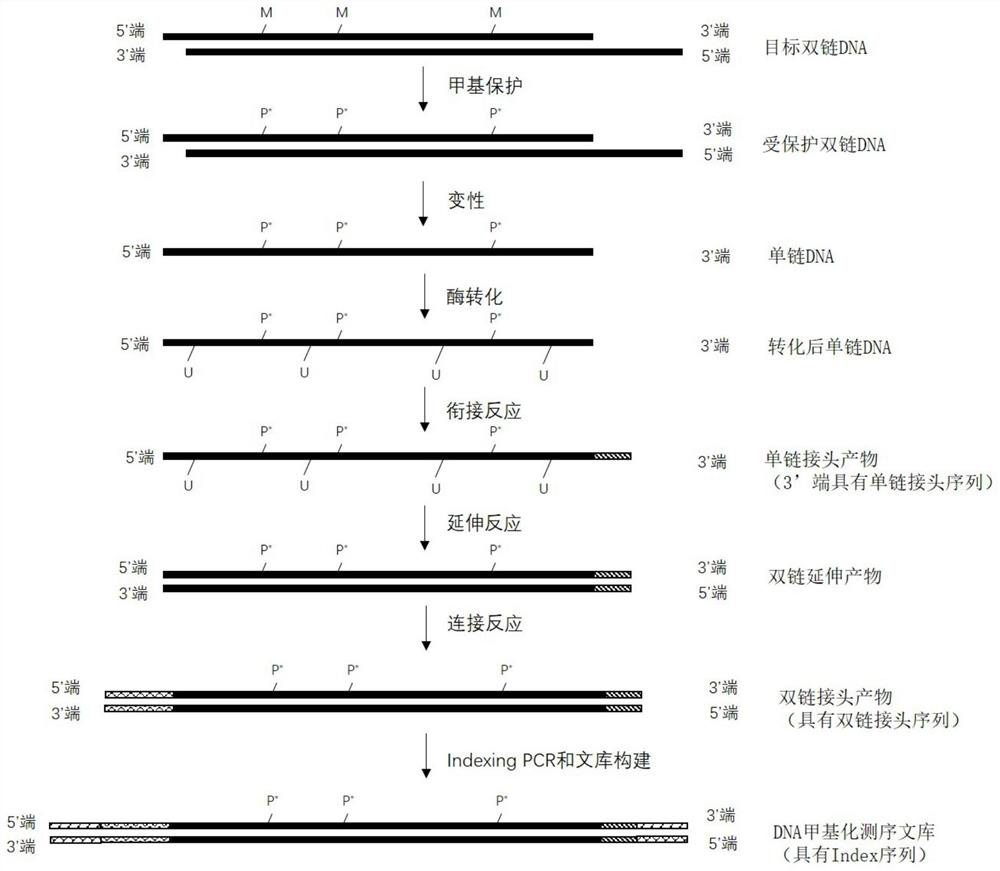
[0091] The present embodiment provides a method for obtaining target double-stranded DNA, which comprises the following steps:
[0092] 1. Extract the DNA in the target sample, detect the concentration of DNA, and use electrophoresis to detect whether the molecular weight of the extracted DNA band belongs to the molecular weight of the target DNA. If so, proceed to the next step. If not, the DNA was re-extracted.
[0093] 2. Ultrasonic fragmentation of extracted DNA, which specifically includes: determining the amount of DNA loaded according to the concentration of DNA and electrophoretic bands. Generally, the initial amount of fragmentation is 150ng, and the total volume is 100μL. Place it in a fragmentation instrument for fragmentation. For fragmentation, the fragmented DNA is subjected to 2% agarose gel electrophoresis detection, and when the DNA fragment is between 200bp and 300bp, the DNA sequencing sample containing the target double-stranded DNA is obtained. The progr...
Embodiment 2
[0097] This embodiment provides a method for methylation protection of target double-stranded DNA, which includes the following steps:
[0098] 1. Add internal reference DNA:
[0099] Take 26 μL of the fragmented DNA sequencing sample obtained in Example 1, add 1 μL of fragmented λDNA and 1 μL of fragmented pUC19 DNA (containing 0.01-0.02ng), and mix to obtain a DNA mixed sample with a total volume of 28 μL.
[0100] In this example, the added pUC19 DNA is fully methylated, that is, all cytosine (C) on it is fully methylated, and the methylation rate of pUC19 is used to indicate the degree of methylation of cytosine in the experiment. protection is effective. If in the final analysis results, the methylation rate of pUC19 is greater than 98%, it can be considered as a successful methylation protection. Therefore, the added pUC19 DNA can be used as an internal control.
[0101] In this example, the added λDNA is completely unmethylated, that is, all the cytosines (C) thereon...
Embodiment 3
[0113] Embodiment provides a kind of method of DNA denaturation, it comprises the steps:
[0114] Take 16 μL of the purified DNA obtained in Example 2 and place it in a PCR reaction tube, add 4 μL of formamide, put it into a preheated PCR instrument, cover with a thermal lid, and incubate at 85°C for 10 minutes. After the incubation, remove it immediately. Store the reaction tubes on an ice box.
[0115] Wherein, formamide is used as a denaturant to denature purified double-stranded DNA into single-stranded DNA. In this application, the methylated cytosine is protected by an oxidation reaction, and then denatured into a single-chain conversion unmethylated cytosine, which is determined by the design of the enzymatic conversion method of NEB Company.
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com