Method for screening CTL epitopes from autonomously constructed SLA-2-HB01-pCDH/sT2 cell line
A technology of sla-2-hb01-pcdh and sla-2-hb01-flag-pcdh, applied in the field of biomedicine, can solve the problems of low efficiency, difficult folding, harsh experimental conditions and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
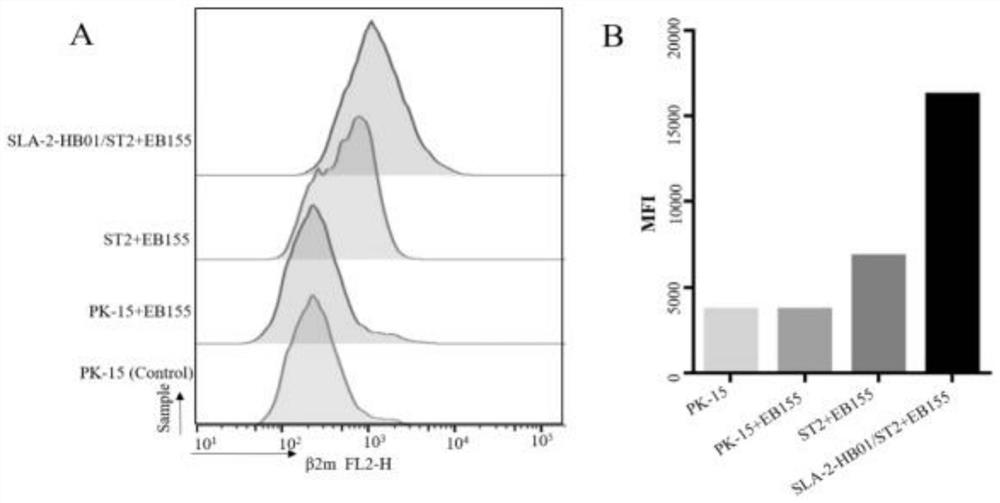
[0091] Example 1 Affinity experiment of PK15, sT2 and SLA-2-HB01-pCDH / sT2 cell lines
[0092] EB155 peptides were used as positive control peptides in PK15, sT2 and SLA-2-HB01-pCDH / sT2 cell lines, and loaded in the three cell lines respectively, because in the previous Western blotting test, the expression of β2m in the cell lines was basically the same , so the porcine β2m monoclonal antibody was selected as the detection of cell surface peptides. Because SLA-I-peptides are presented to the surface of the cell membrane as a whole, the detection of the expression of β2m on the surface of the cell membrane indicates that there are epitope peptides presented to the cell surface by SLA class I molecules. In the experimental group, only PE-labeled IgG was set as the background control group, which was no different from pure PK15 and sT2 cells, indicating that the staining result was the result of flow cytometry detection of the specific combination of PE and β2m monoclonal antibod...
Embodiment 3
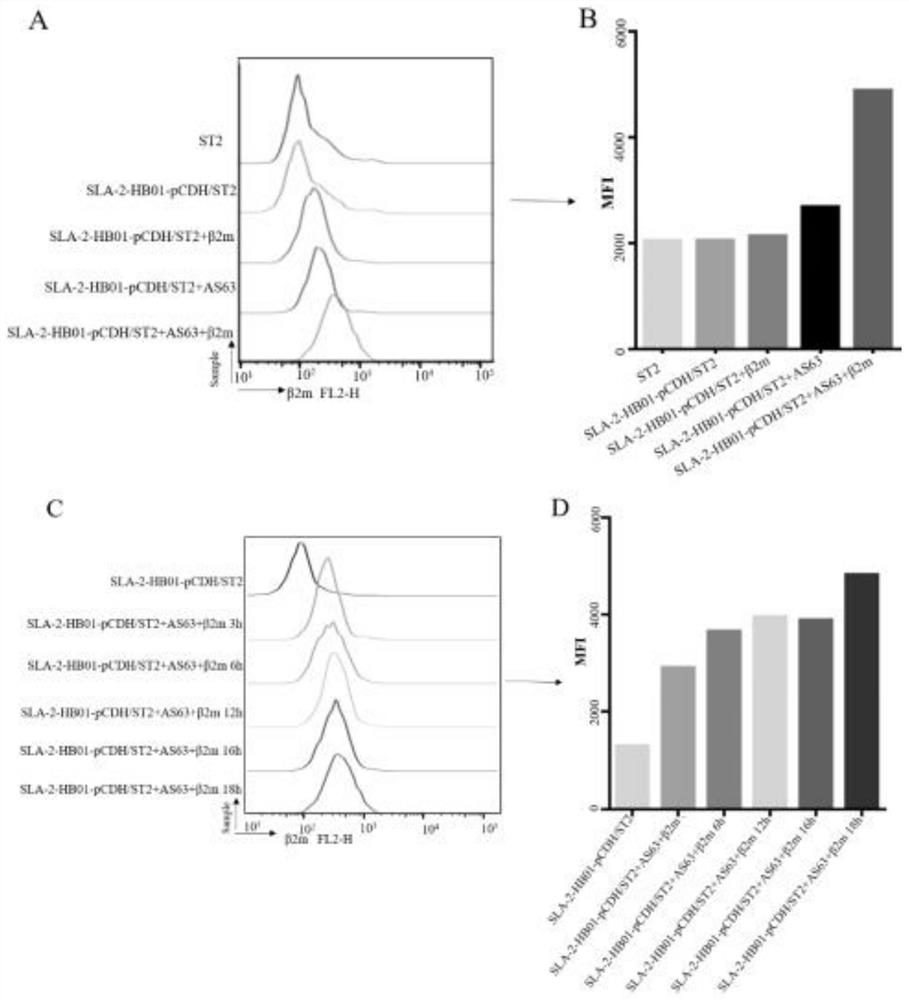
[0095] Example 3 CTL epitope screening function verification
[0096] After determining the peptide screening conditions, use the binding experiment of peptides loaded on SLA-2-HB01-pCDH / sT2 cells to detect whether the peptides can bind to SLA-2-HB01 and whether the affinity is enhanced, 50 μg / mL exogenous Peptides and 3 μg / mL β2m were added to the above cell lines, incubated at 37°C for 18 hours, and then the expression of β2m on the cell membrane surface was measured by β2m monoclonal antibody, so as to determine the expression of SLA-2-peptides and screen out CTL epitope peptides, such as Figure 4 shown. The mean fluorescence intensity (MFI) of β2m on the cell lines was determined by FACs analysis, and the fluorescence index (FI) was calculated. According to the following formula: FI=(MFI[SLA-2-HB01-pCDHsT2 cells+peptide] / MFI[SLA-2-HB01-pCDHsT2 cells without peptide])-1 proves the binding ability of SLA-2 to peptides. The results showed that AS63, Hu62 and EB155 could al...
Embodiment 4
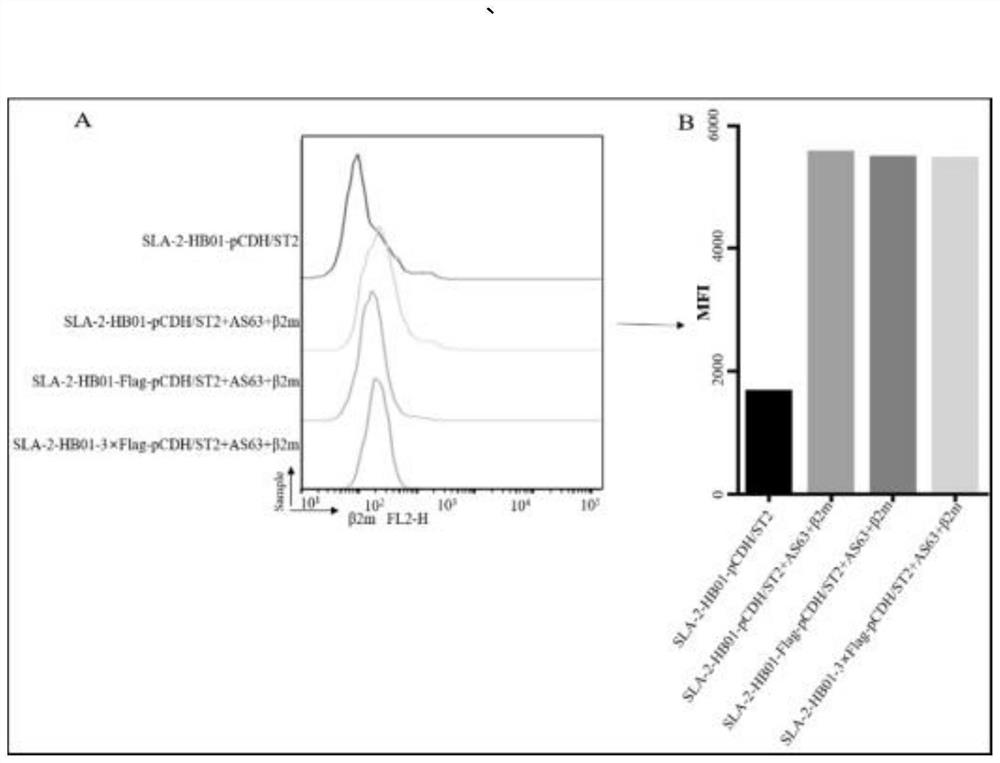
[0099] Example 4 Laser confocal experiment to verify cell line function
[0100] Add 100 μg / mL Biotin-AS63 peptide and 3 μg / mL β2m to the laser confocal plate of SLA-2-HB01-Flag-pCDH / sT2, SLA-2-HB01-3×Flag-pCDH / sT2 cells and incubate at 37°C for 18 hours Finally, the SLA I-peptide-β2m complex was stained with trimolecular cells, and the specific staining structure was as follows Figure 5 . LSCM detection results show that in the wavelength range of 590-617nm, after loading Biotin-AS63 peptides in SLA-2-HB01-Flag-pCDH / sT2 cells, there is no Alexa594-anti Flag antibody on the surface (1:50 dilution) Fluorescence expressed after binding the Flag tag see Figure 6 B, but in the SLA-2-HB01-3×Flag-pCDH / sT2 cell line under the same conditions, Alexa594-anti Flag antibody (diluted 1:50) was observed to bind to the corresponding 3×Flag fluorescence on the cell membrane surface as Figure 6 d.
[0101] In the wavelength range of 400-420nm, SLA-2-HB01-Flag-pCDH / sT2 and SLA-2-HB01-3×...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com