Specific primer and probe combination for qPCR (quantitative polymerase chain reaction) quantitative detection of copy number of target gene of urothelial carcinoma and application of specific primer and probe combination
A technology for urothelial cancer and quantitative detection, which is applied in the determination/inspection of microorganisms, recombinant DNA technology, biochemical equipment and methods, etc., and can solve the problems of complicated operation, long detection time, and poor specificity of detection methods.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
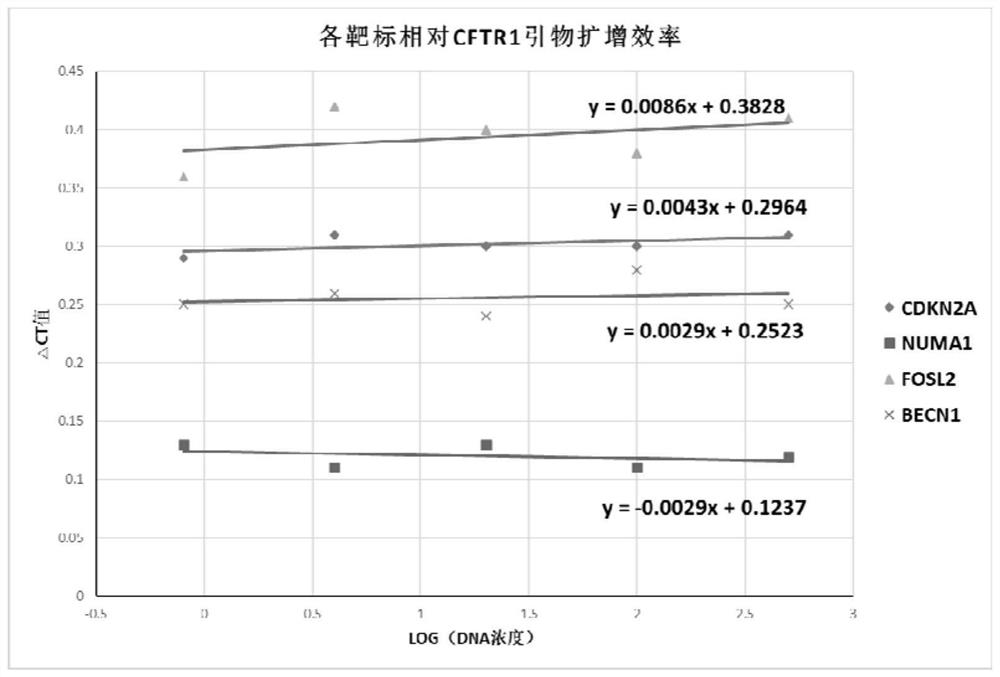
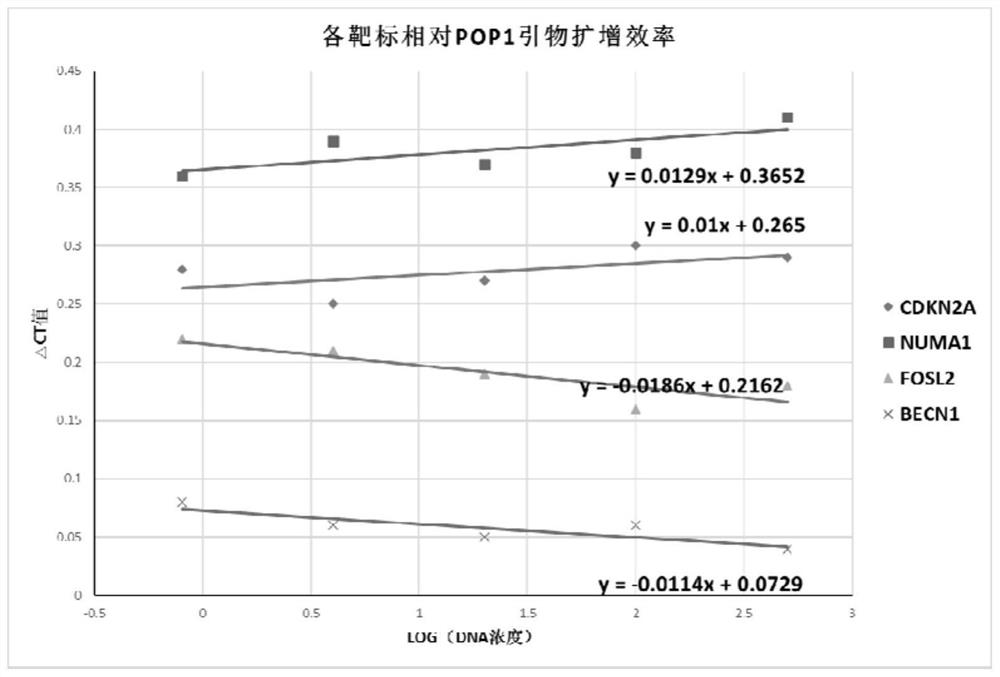
[0061] Assess primer amplification efficiency for target genes and internal reference genes
[0062] The specific primers and probe sequences of CDKN2A gene are shown in Table 1:
[0063] Table 1
[0064] name sequence(5'->3') serial number forward primer TCCCAGTCTGCAGTTAAGGG SEQ ID NO.1 reverse primer GGAGGGTCACCAAGAACCTG SEQ ID NO.2 detection probe CCTCTGGTGCCAAAGGGCGG SEQ ID NO.3
[0065] The specific primers and probe sequences of NUMA1 gene are shown in Table 2:
[0066] Table 2
[0067] name sequence(5'->3') serial number forward primer AAGACTGAAATCCTAACAGGGCAG SEQ ID NO.4 reverse primer CAGGTCAACGGGGTGAGTCAG SEQ ID NO.5 detection probe GTGTGGGTGTGGCTTGGCAGT SEQ ID NO.6
[0068] The specific primers and probe sequences of FOSL2 gene are shown in Table 3:
[0069] table 3
[0070] name sequence(5'->3') serial number forward primer CCTGGAGGGAAGTCAGACCG SEQ ...
Embodiment 2
[0093] Detection of gene copy number
[0094] The CDKN2A gene reaction solution was prepared, wherein the CDKN2A gene forward and reverse primer concentrations were 20 μM, the probe concentration was 5 μM, and the forward and reverse primer concentrations of the internal reference genes CFTR1 gene and POP1 gene were both 5 μM;
[0095] Prepare a NUMA1 gene reaction solution, wherein the concentration of the forward and reverse primers of the NUMA1 gene is 20 μM, the concentration of the probe is 5 μM, and the concentrations of the forward and reverse primers of the internal reference genes CFTR1 gene and POP1 gene are both 5 μM;
[0096] Prepare a FOSL2 gene reaction solution, wherein the concentration of the forward and reverse primers of the FOSL2 gene is 20 μM, the concentration of the probe is 5 μM, and the concentrations of the forward and reverse primers of the internal reference genes CFTR1 gene and POP1 gene are both 5 μM;
[0097] Prepare the BECN1 gene reaction solut...
Embodiment 3
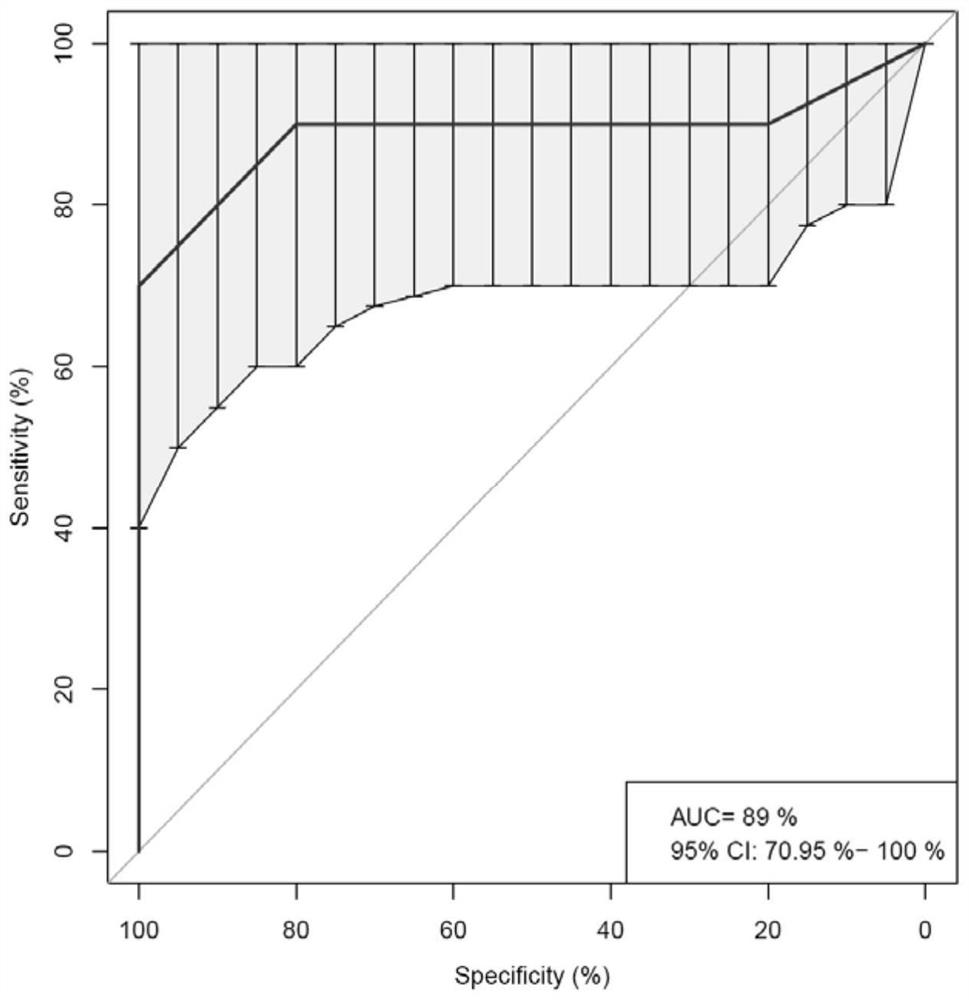
[0117] Establishment of predictive model for auxiliary diagnosis of urothelial carcinoma
[0118] Collect 50 urothelial carcinoma-positive and 30 urothelial-carcinoma-negative samples respectively. All samples come from hospitals, and the judgment of positive and negative is based on clinical diagnosis. Take 30mL urine sample to separate urine precipitate and extract DNA.
[0119] The CDKN2A gene reaction solution was prepared, wherein the CDKN2A gene forward and reverse primer concentrations were 20 μM, the probe concentration was 5 μM, and the forward and reverse primer concentrations of the internal reference genes CFTR1 gene and POP1 gene were both 5 μM;
[0120] Prepare a NUMA1 gene reaction solution, wherein the concentration of the forward and reverse primers of the NUMA1 gene is 20 μM, the concentration of the probe is 5 μM, and the concentrations of the forward and reverse primers of the internal reference genes CFTR1 gene and POP1 gene are both 5 μM;
[0121] Prepar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com