Method and device for correcting homologous recombination repair defect score
A homologous recombination and defect technology, applied in the field of bioinformatics, can solve the problems of limited application, high sequencing data volume, inability to cover somatic cell mutations, etc., and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
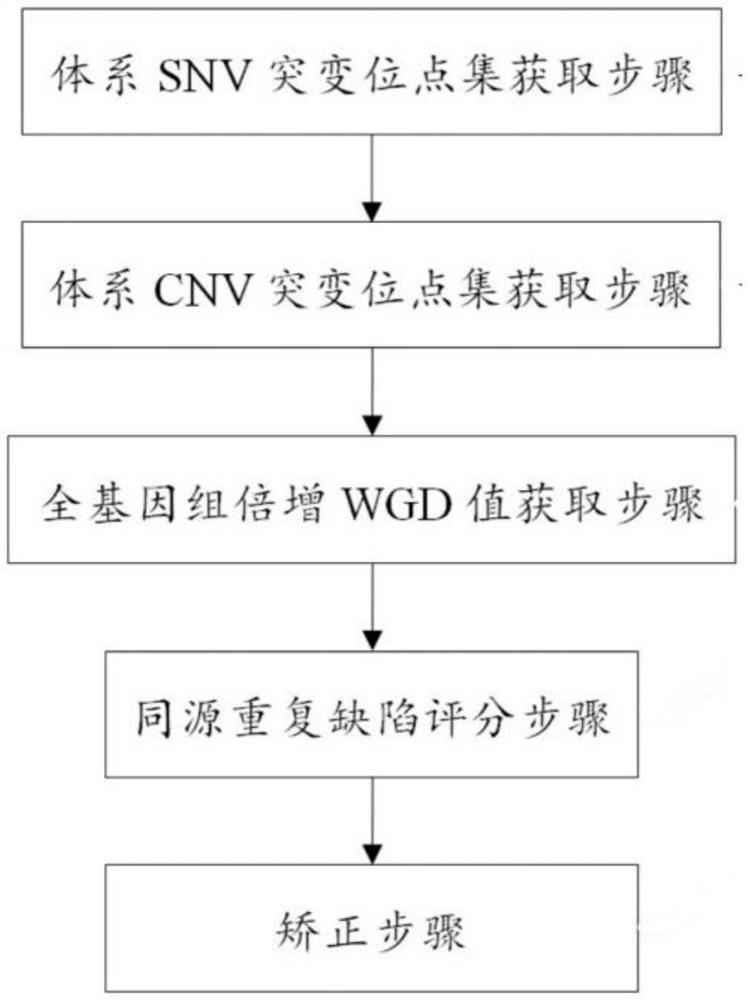
[0092] The evaluation method of this embodiment mainly includes the following steps:
[0093] (1) Sequencing data files (bam format) generated after steps such as comparison, sorting, filtering, and labeling and repetition using sequencing off-machine data for tumor samples and paired blood cell samples;
[0094] (2) Use Mutect2 software to analyze the SNV of the tumor samples to be tested, and obtain the mutation set of the system SNV, including information such as mutation frequency and mutation site depth;
[0095] (3) Annotate the obtained system SNV;
[0096] (4) Use the cnvkit software to take the bam of the tumor sample to be tested and the paired sample as input, analyze and obtain the segment of the CNV mutation in the sample, and output the size of the segment, the number of probes contained in the segment, and the segment BAF value and other information;
[0097] (5) Use the ABSOLUTE software to analyze the purity and ploidy of the tumor samples to be tested, and ...
Embodiment 2
[0218] Embodiment 2 HRD accuracy evaluation
[0219] In this embodiment, the method in Example 1 (abbreviated as WES detection method) is used for evaluation.
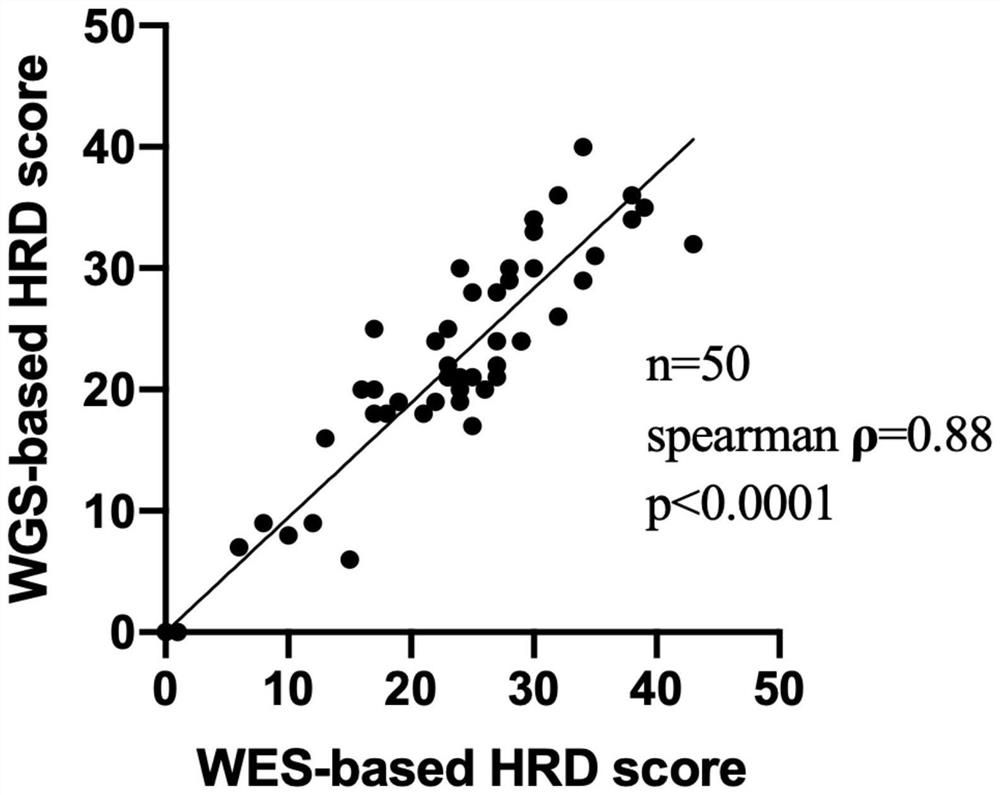
[0220] (1) Consistency comparison with WGS detection
[0221] In this embodiment, 50 ovarian cancer FFPE samples (formalin-fixed and paraffin-embedded samples, Formalin-Fixed and Parrffin-Embedded, referred to as FFPE samples) were used, and the results detected by the WES detection method based on Example 1 were compared with Compared with the HRD values obtained by WGS detection, the results are as follows figure 2 As shown, Spearman ρ=0.88, showing a good consistency.
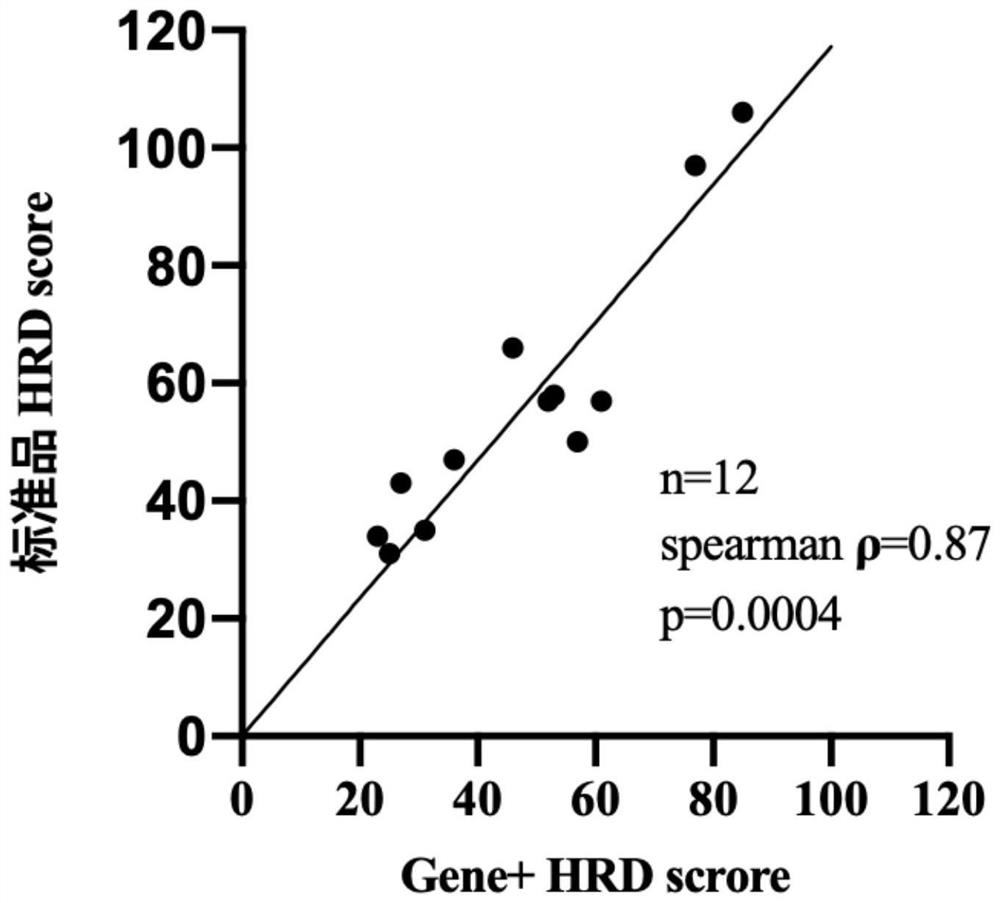
[0222] (2) Consistency comparison with the standard
[0223] The present embodiment has detected 12 pairs of HRD standard items, by comparing with reference value, investigates the accuracy of this detection method, the result is as follows image 3 It shows that the HRD detection result of the detection method of this embodiment has a high co...
Embodiment 3
[0224] Embodiment 3HRD score and clinical curative effect
[0225] In this example, the method of Example 1 was used to detect the samples of 59 patients with high-grade serous ovarian cancer treated with platinum-based chemotherapy drugs for the first time, and the relationship between the first platinum curative effect data and HRD was calculated, including 45 patients with platinum-sensitive Data and 14 cases of platinum resistance data. Statistics see Figure 4 . from Figure 4 It can be seen that HRD-positive samples have a higher PFS than HRD-negative samples (Log rank p=0.04, HR=0.55). PFS refers to the progression-free survival period (progression-free survival). Time to disease progression or death from any cause.
[0226] Those skilled in the art can understand that all or part of the functions of the various methods in the foregoing implementation manners can be realized by means of hardware, or by means of computer programs. When all or part of the functions i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com