Homologous recombination defect gene analysis method
A gene analysis and homologous recombination technology, applied in the field of gene analysis, can solve the problems of difficult HRD status assessment, high cost, and technical complexity, and achieve the effects of improving the economic efficiency of detection, reducing detection cost, and expanding the detection rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
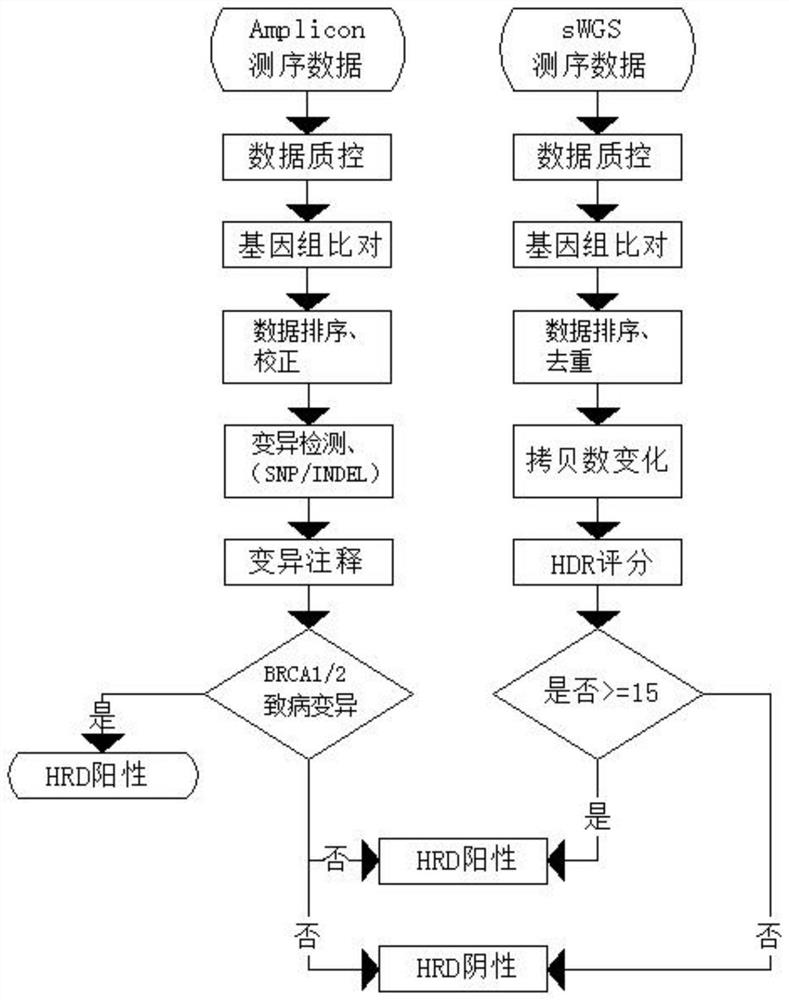
[0046] Based on the results of the above two steps, if the HRD score is greater than or equal to 15 in the first step, and there is no pathogenic variant in the BRCA1 / 2 gene in the second step, it is judged to be HRD positive.
Embodiment 2
[0048] Based on the results of the above two steps, if the HRD score is judged to be less than 15 in the first step, and there is no pathogenic variant in the BRCA1 / 2 gene in the second step, it is judged to be HRD negative.
Embodiment 3
[0050] Based on the results of the above two steps, if there is a pathogenic / possible pathogenic variant in the BRCA1 / 2 gene, it is judged to be HRD positive.
[0051] To sum up, with reference to the data, this method adopts the low-depth whole genome sequencing (sWGS) technology with a sequencing depth of about 1X, and directly and rapidly detects HRD status by mining copy number changes; combined with BRCA1 / 2 gene variation Whether or not, it has achieved a comprehensive assessment of homologous recombination repair defects, and comprehensively judged whether it benefits from the treatment of PARP inhibitors. This method can accurately evaluate genome instability, which can effectively expand the positive detection rate of homologous recombination defects, and Reduce detection costs; at the same time, it has the advantages of less time-consuming and can be used for single-sample analysis. This method combines BRCA1 / 2 gene variation; at the same time, it meets the in-depth r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com