Composition and method for determining enzyme activity of M-MLV reverse transcriptase
A technology of reverse transcriptase and assay method, which is applied in the field of M-MLV reverse transcriptase enzyme activity assay composition, can solve the problems of large human factors, aerosol pollution, and long time consumption, and achieve simple assay operation and good repeatability , the effect of simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0135] SYBR dye relative enzyme activity determination process:
[0136] 1. Dilute different commercial enzymes, take 1ul each time and mix with the Mix in step 2.
[0137]
[0138] 2. Mix preparation
[0139]
[0140]
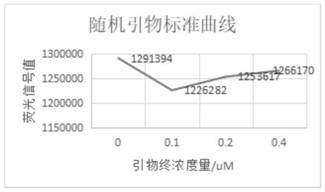
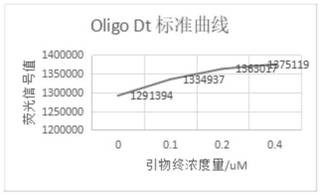
[0141] 4. Data processing, see Figure 5 .
[0142]
[0143] 4. Experimental analysis
[0144] SYBR dye tests the enzyme activity of M-MLV enzyme, can establish standard curve and R 2 Can reach 0.99
Embodiment 2
[0146] 1. Dilute Oligreen dye to 1X
[0147] 2. RNase A enzyme preparation
[0148] (1) Weigh 100mg of RNase A powder (using Gao Bingbing's balance), and put it into a 50ml centrifuge tube;
[0149] (2) Add 100uL Tris-HCl(1M), 50uLNaCl(3M), add 7mlNF water to dissolve RNase A powder;
[0150] (3) Adjust the pH value of the solution to 7.5 with HCl and NaOH, then dilute to 10ml with water, and mix the solution thoroughly;
[0151] (4) Boil in a metal bath at 100°C for 15 minutes, and slowly drop to room temperature (flocculent formation occurs);
[0152] (5) Centrifuge the RNase A solution cooled to room temperature at 3500rmp for 5min, filter it with a 0.22um filter element, and store it at -20°C.
[0153] 3. Mix preparation
[0154] TE addition amount / uL 9.27 9.02 8.77 8.27 7.77 293T-RNA amount / uL 0.73 0.73 0.73 0.73 0.73 RNase A enzyme amount / uL 0 0.25 0.5 1 1.5 Oligreen amount / uL 10 10 10 10 10
[0155] 4. React on a mic...
Embodiment 3
[0164] 1. Dilute Oligreen dye to 1X
[0165] 2. Commercial enzyme (Maxima H Minus Reverse Transcriptase) dilution
[0166]
[0167] 3. Mix preparation
[0168]
[0169]
[0170] 4. Perform amplification on the microplate reader and instrument, (42°C, 1min) for 20min
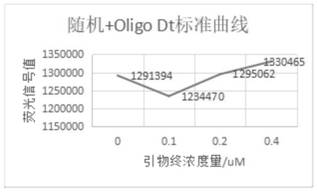
[0171] 5. Experimental map, see in details Figure 8
[0172] 6. For data processing, see Figure 9
[0173]
[0174] 7. Experimental analysis
[0175] (1) There is a background signal in the system, and when the amount of enzyme added is 0, Oligreen may combine with single strands such as RNA and primers to produce fluorescence;
[0176] (2) The more MMLV enzyme added in the system, the lower the fluorescence signal;
[0177] (3) When Oligreen dye detects M-MLV enzyme, there is a change in fluorescence signal, which can be used to test the enzyme activity.
[0178] It should be noted that in this article, relational terms such as first and second are only used to distinguish one entity or ope...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com