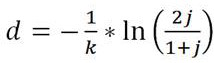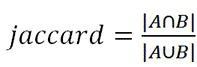Large-scale biological data clustering method and system based on spanning tree
A technology of biological data and clustering method, applied in the field of data processing, can solve the problems of not being able to fully utilize and exert the advantages of parallel computing of multi-core platforms, and achieve the effect of avoiding storage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
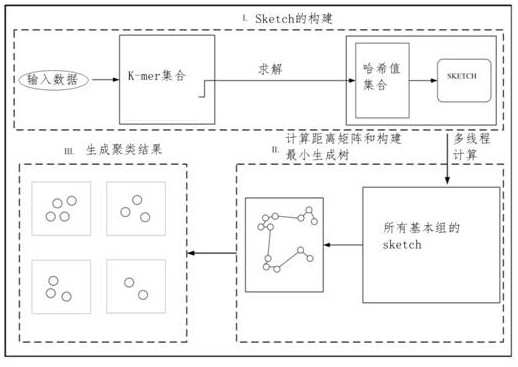
[0043] This embodiment discloses a large-scale biological data clustering method, including:
[0044] Step 1: Construction of Sketch. This step is mainly to generate k-mers from the original biological genome sequence through a sliding window to obtain a set of k-mers, and then map the k-mers in the set into corresponding hash values through a hash function, and select them by the minHash method Among them, the smallest fixed number (1000) of hash values is saved as a sketch, and this fixed number is the dimension of the sketch. Since the hash function that maps k-mer to hash value satisfies uniformity, that is, the hash value mapped to k-mer is equally distributed in the corresponding hash value space, so the selected minimum fixed number The hash value of the number is equivalent to a fixed number of k-mers randomly selected from all k-mers. And all genome sequences use the same hash function, which ensures that the same k-mer input has the same hash value output, so t...
Embodiment 2
[0062] The purpose of this embodiment is to provide a large-scale biological data clustering system, including:
[0063] A similarity estimation module for estimating the similarity between genome sequences;
[0064] The minimum spanning tree generation module is used to calculate the distance matrix between the genome sequences and construct the minimum spanning tree based on the similarity between the estimated genome sequences, and generate the minimum spanning tree by dividing the distance matrix into subgraphs and constructing the sub-minimum spanning tree Tree;
[0065] The clustering module is used to prune edges exceeding a given threshold length in the minimum spanning tree to generate clustering results.
[0066] Those skilled in the art should understand that each module or each step of the present invention described above can be realized by a general-purpose computer device, optionally, they can be realized by a program code executable by the computing device, th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com