SNP (Single Nucleotide Polymorphism) molecular marker related to growth traits of black sea bream and application of SNP molecular marker
A technology of molecular markers and growth traits, which is applied in the field of molecular biology, can solve the problems of black sea bream parent quality, large growth disparity, cannibalism and other problems, so as to shorten the breeding cycle, speed up the breeding process and improve the accuracy Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
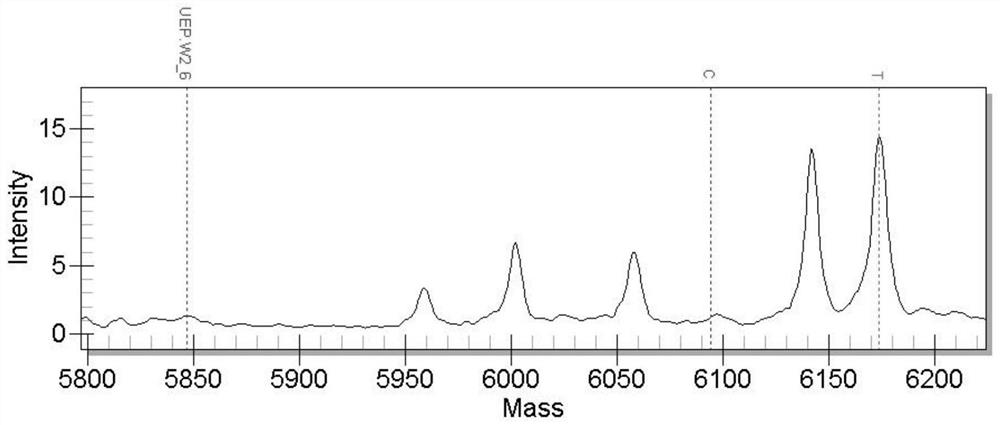
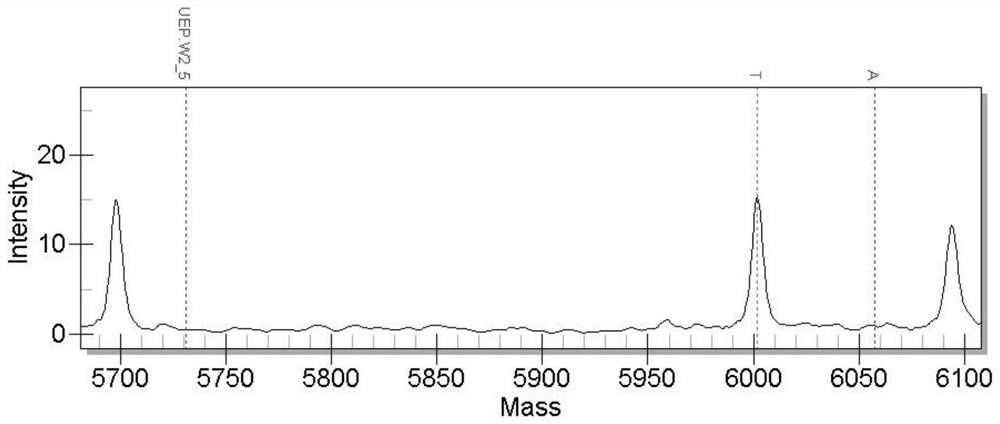
[0029] 1. Acquisition of SNP molecular markers
[0030]The experimental fish used were the same batch of newly hatched black sea bream, cultivated in outdoor ponds, and fed with feed twice a day. During the cultivation period, the water temperature and salinity were within the adaptable range of black sea bream cultivation. At 4 months of age, the survival rate of black sea bream is 100%. Select 5 extremely large individuals (fast growth) and 5 extremely small individuals (slow growth) from all black sea breams, cut tissue samples immediately after anesthesia, then extract the total RNA of each fish, and take an equal amount of total RNA The RNA libraries of the fast-growing group and the slow-growing group were constructed, and transcriptome sequencing was performed. The sequencing results were compared with the whole genome sequence of black sea bream. After analysis by bioinformatics software, a large number of potential SNP sites were obtained, among which adenylate kinas...
Embodiment 2
[0044] Utilize the SNP molecular marker that embodiment 1 screens out to screen the black sea bream parent with high growth rate, comprise the steps:
[0045] (1) Cut the black sea bream caudal fin sample to be tested, extract DNA;
[0046] (2) Using the extracted DNA as a template, the primer pair is used to perform PCR amplification to obtain the primary PCR amplification product;
[0047] Among them, the primer pair for detecting the SNP molecular marker located in the adenylate kinase 2 gene is:
[0048] F1: 3'-ACGTTGGATGTCGTGGAGCTCATAGAGAAG-5' (SEQ ID NO.4)
[0049] R1: 3'-ACGTTGGATGTCTCCGCTTGTTTGACTGTG-5' (SEQ ID NO.5)
[0050] Extension primer 1: 3'-AGAAGAACTTGGACACGTC-5' (SEQ ID NO.6);
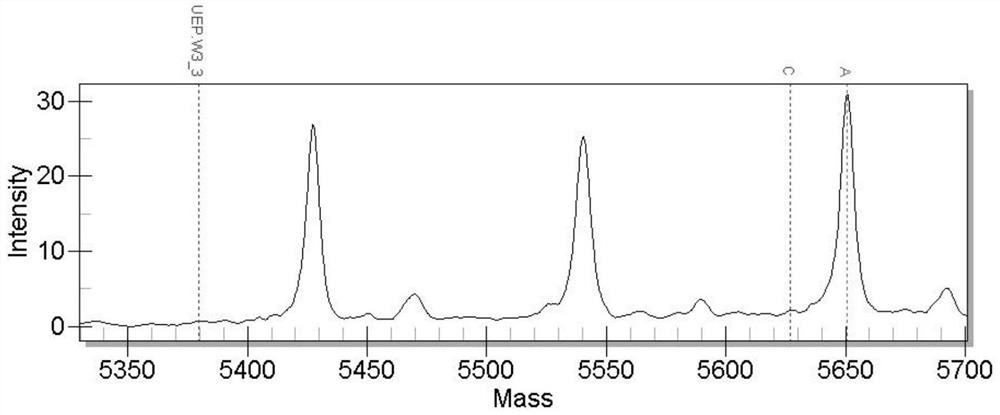
[0051] The primer pair used to detect the SNP molecular marker located in the cytochrome P4503A gene is:
[0052] F2: 3'-ACGTTGGATGTACCCTTTGGAACCACAAGC-5' (SEQ ID NO.7)
[0053] R2: 3'-ACGTTGGGATGTTTGTACCCCATCCTTCCAC-5' (SEQ ID NO.8)
[0054] Extension primer 2: 3'-CACAAGCCCATTTA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com