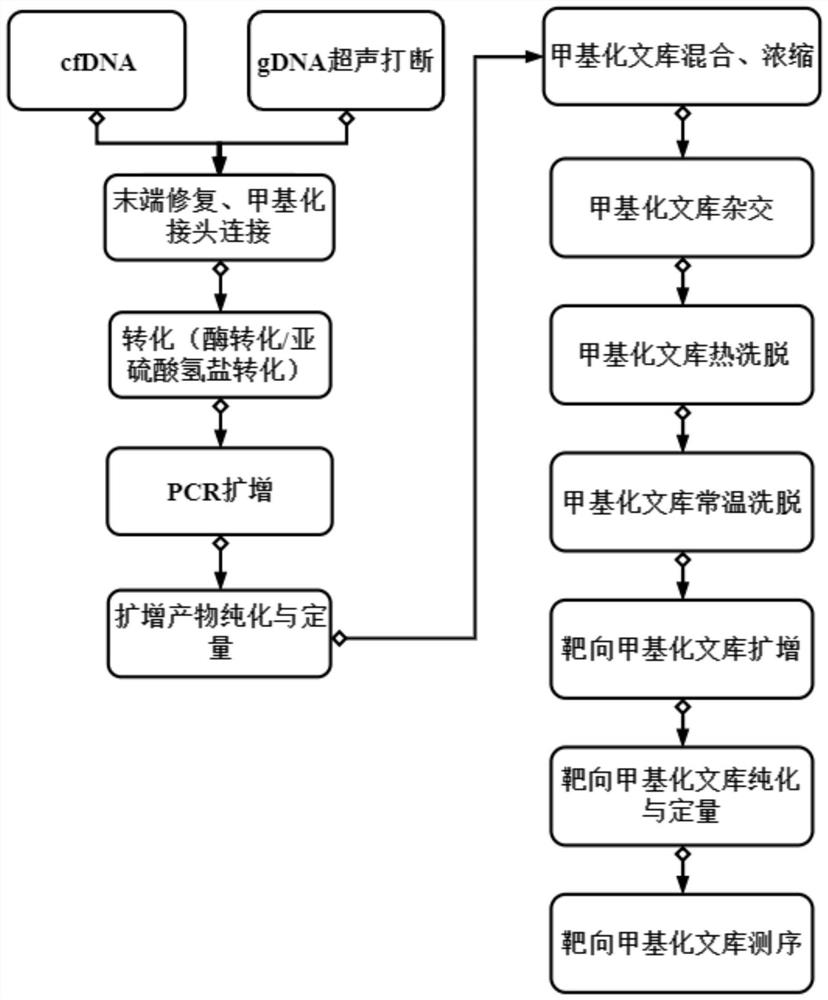
Construction method of targeted methylation sequencing library and kit
A construction method and technology for sequencing libraries, which are applied in the field of construction methods and kits for targeted methylation sequencing libraries, can solve the problems of low template requirements, low data capture preference, and high template input requirements, and achieve improved Capture efficiency, optimized hybridization capture temperature, and high library diversity effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Different thermal elution reagent tests
[0035] Step 1: Library hybridization
[0036] 1. Preparations
[0037] Take out the hybridization reagent 2X Hyb Buffer and Hyb 2 in the hybridization capture kit in advance and dissolve at room temperature; note that 2X Hyb Buffer must be fully dissolved until completely free of crystals. If it cannot be dissolved at room temperature, it can be incubated in a water bath at 65°C, and vortexed during the mixing , until the crystals are completely dissolved; after the first use, a small amount of packaging is convenient for the next dissolution and use. Turn on the power of the vacuum concentrator, adjust the temperature to 45°C, and preheat in advance.
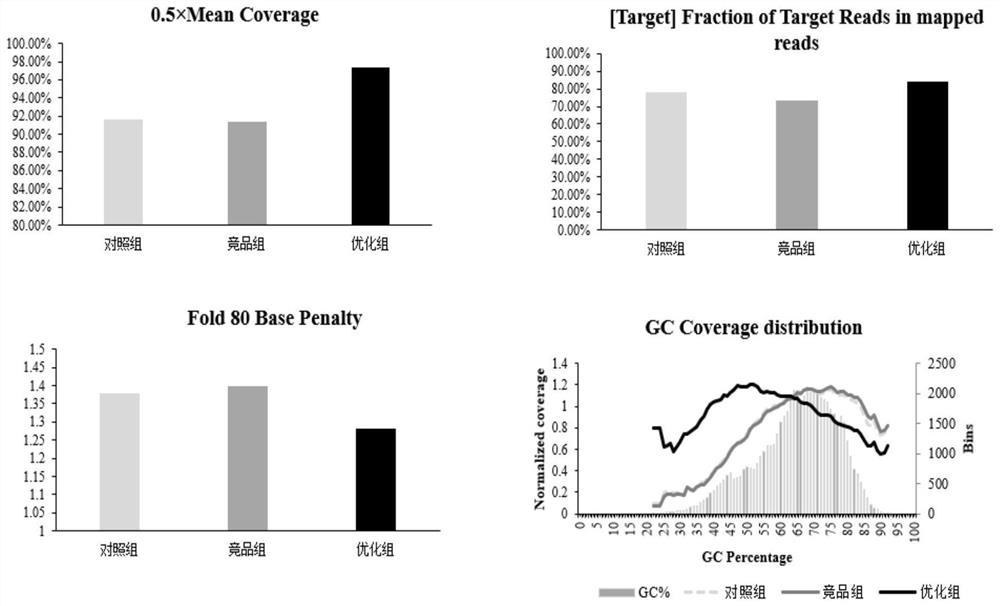
[0038] 2. Experimental grouping, gDNA (Human Genomic DNA: Female; G1521, PromegaCovaris interrupted to 200-250bp) after ultrasonic fragmentation was combined with the EM transformation module to construct a methylation library, and the following three methods were used to constr...
Embodiment 2
[0112] Enzyme Conversion Libraries Using Different Hybridization Capture Temperatures
[0113] Step 1: Library hybridization
[0114] 1. Preparations
[0115] Take out the hybridization reagent 2X Hyb Buffer and Hyb 2 in the hybridization capture kit in advance and dissolve at room temperature; note that 2X Hyb Buffer must be fully dissolved until completely free of crystals. If it cannot be dissolved at room temperature, it can be incubated in a water bath at 65°C, and vortexed during the mixing , until the crystals are completely dissolved; after the first use, a small amount of packaging is convenient for the next dissolution and use. Turn on the power of the vacuum concentrator, adjust the temperature to 45°C, and preheat in advance.
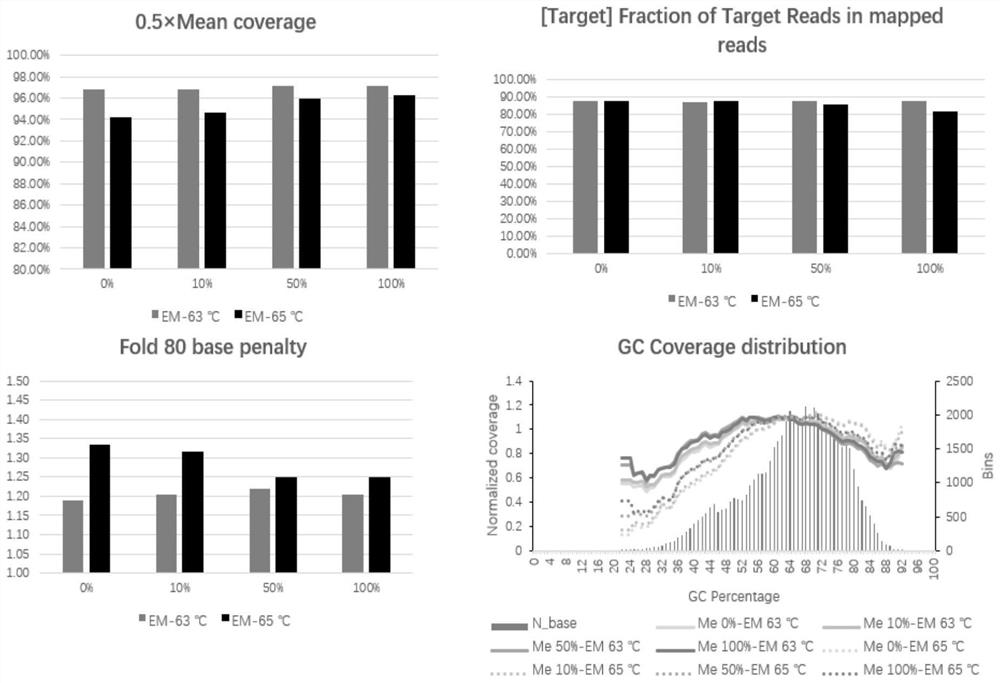
[0116] 2. Experimental grouping, methylation standard zymo, Methylated DNA, D5014-2 and zymo, Non-Methylated DNA, D5014-1 were mixed according to a certain ratio to form a methylation level of 0%, 10%, 50% and The 100% methylation standard...
Embodiment 3
[0151] Bisulfite-Converted Libraries Using Different Hybridization Capture Temperatures
[0152] Step 1: Library hybridization
[0153] 1. Preparations
[0154]Take out the hybridization reagents 2X Hyb Buffer and Hyb 2 in the hybridization capture kit in advance and dissolve them at room temperature; note that the 2X Hyb Buffer must be fully dissolved until completely free of crystals. Rotate and mix until the crystals are completely dissolved; after the first use, aliquot a small amount to facilitate the next dissolution and use. Turn on the power of the vacuum concentrator, adjust the temperature to 45°C, and preheat in advance.
[0155] 2. Experimental grouping, using gDNA fragmented to 200-250bp, with Qiagen's bisulfite conversion module (QIAGEN Fast DNA Bisulfite Kit (Cat#59824 / 59826)), construct a methylation library, and use the three schemes shown in Table 8 in Example 2 to construct a targeted methylation library.
[0156] 3. Library Mixing
[0157] See Example ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com