Primer for fluorescent quantitative detection of strawberry mottle virus and application of primer
A fluorescent quantitative detection, mottled virus technology, applied in the biological field
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1, determination of strawberry mottle virus fluorescence quantitative detection method
[0028] First, the determination of primers
[0029] The inventors of the present invention in the study of strawberry mottled virus and sequence determination, screened and synthesized the following SMoV fluorescence quantitative detection primer sequence, SM6364F: 5′CGCCGACAGTTCTCTATG3' (sequence in the sequence list 1), SM6517R: 5'ATACACCACCAGCGCCTCTTT3 '(sequence 2 in the sequence list). The primers are synthesized in Shanghai Shenggong Company.
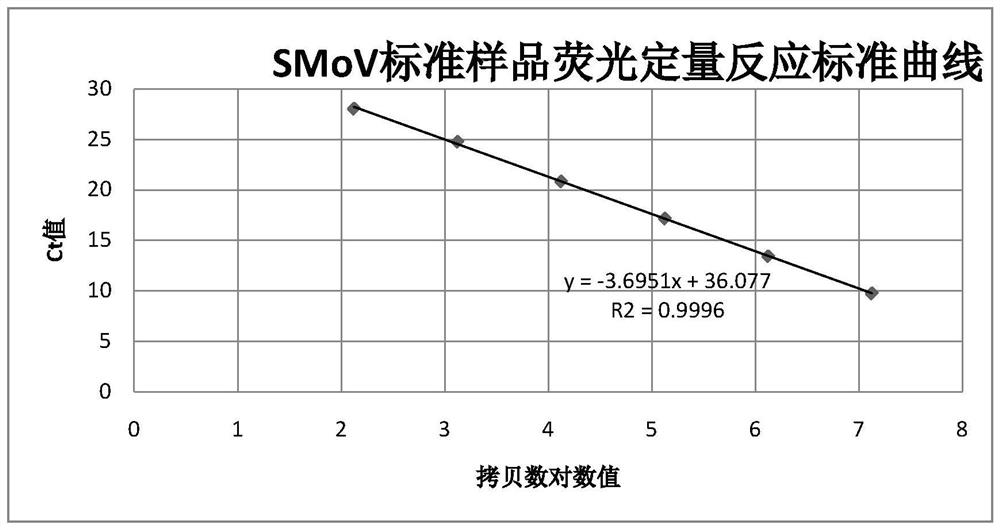
[0030] Second, the establishment of standard curves
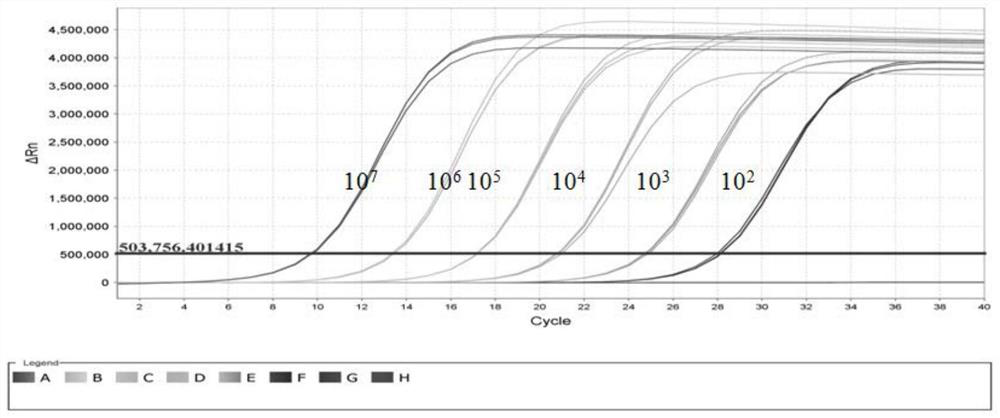
[0031] 1. Amplification of the target fragment
[0032] The RNA extracted from the SMoV-positive strawberry leaf samples collected in 2019.10 in Changping Jinliuhuan, Beijing was reverse-recorded into cDNA, and the SMoV target fragment was amplified using primers SM6364F: CGCCGACAGTTCTCTATG, SM6517: ATACACCACCAGCCTCTT, with a fragment size of 153 bp and a target fragment located ...
Embodiment 2
[0054] Example 2, real-time fluorescence quantification method for the detection of field samples
[0055] 1. Sample source
[0056] 2019.10 40 strawberry samples collected from Beijing Changping Jinliuhuan Agricultural Park, including SMoV strawberry plants Liaoning Donggang Hongyan No. 3, Liaoning Donggang Hongyan No. 9 and negative samples as control, were placed at -80 °C for storage.
[0057] 2. Detection of field samples
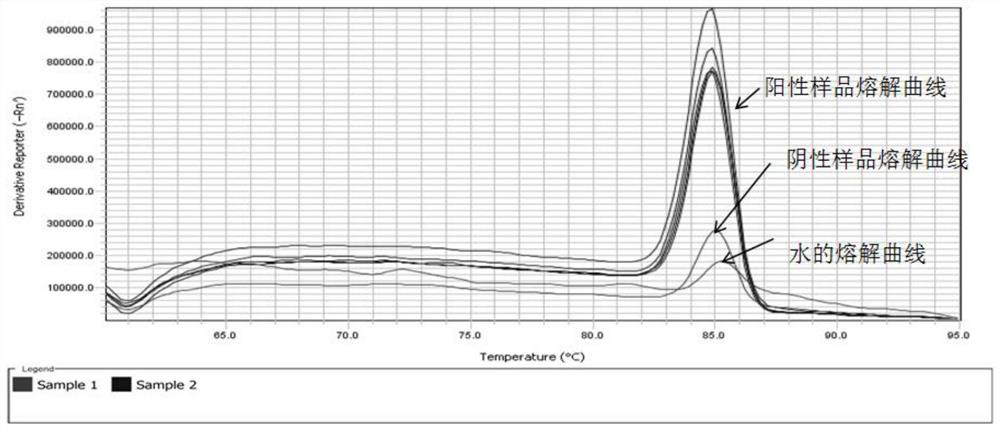
[0058] RT-qPCR and RT-PCR were used to detect whether 40 strawberries were infected with SmoV.
[0059] The RT-qPCR method is as follows:
[0060] (1) Extraction of the total RNA of the biological sample to be measured;
[0061] (2) RNA in step (1) as a template, reverse transcription to cDNA, with the fluorescence quantitative detection primers according to claim 1 for fluorescence quantitative detection;
[0062] (3) Via QuantStudio TM 6Flex fluorescence quantitative thermal cycler instrument for detection, and experimental validity judgment; If there is...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com