Inhibitor for knocking down circXPO1 and application of inhibitor in preparation of glioma treatment medicine
A technology for glioma and glioma cells, which can be used in anti-tumor drugs, drug combinations, medical preparations containing active ingredients, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Sanger sequencing and RNase R treatment confirmed the formation of circular RNA
[0026] 1. Experimental method
[0027] 1. RT-PCR to obtain circXPO1
[0028] Total RNA of GBM cell lines (ie U87-MG, U251 cells) was extracted and reverse transcribed using Trizol reagent (ThermoFisher Scientific) for RT-PCR analysis. Primers are shown in (Table 1).
[0029] RT-PCR system:
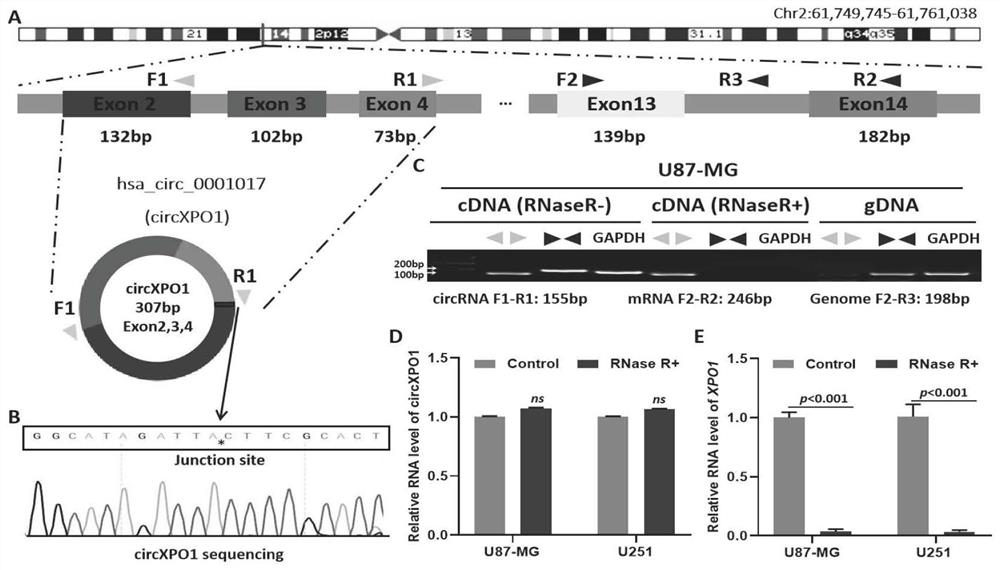
[0030] In order to prove that circXPO1 forms circular RNA rather than linear, the RT-PCR product was recovered and sent to the company for sanger sequencing, and the sequencing results were compared with DNASTAR software. The results show that circXPO1 is indeed formed by the head-to-tail circularization of exons 2, 3, and 4 of the parent gene XPO1 ( figure 1 A, B, C).
[0031] 2. To confirm the circular structure of circXPO1, its stability was assessed by treatment with exonuclease (RNase R). The results showed that circXPO1 was resistant to RNase R ( figure 1 D), while XPO1 was di...
Embodiment 2
[0034] Example 2: Expression of circXPO1 in normal and glioma tissues and GBM cell lines
[0035] 1. Experimental method:
[0036] 1. Fluorescence quantitative PCR to detect the expression of circXPO1 in normal and glioma tissues and GBM cell lines
[0037]Total RNA was extracted from glioma tissue and GBM cell lines (i.e. U87-MG, U251 cells), and Trizol reagent (Thermo Fisher Scientific) was used for quantitative PCR (qRT-PCR) analysis. The amplification reaction was completed by the cfx96 real-time PCR detection system (Bio-Rad Company, Hercules, CA, USA) using SYBR Green fluorescence quantitative PCR kit according to the operating specifications. For circXPO1 and XPO1 primers (Table 1), use The expression of related genes was calculated by the method. All qRT-PCR experiments were repeated in triplicate and all data used GAPDH as a control.
[0038] qRT-PCR system:
[0039] 2. Experimental results
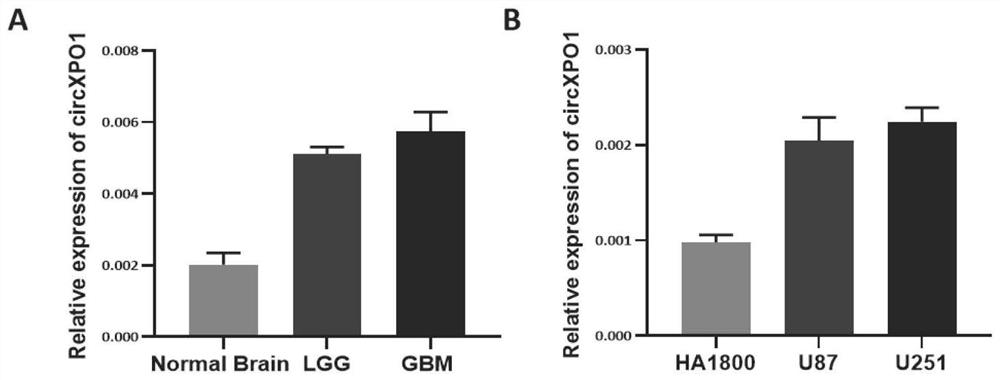
[0040] The expression of circXPO1 in normal brain tissue, low-grade...
Embodiment 3
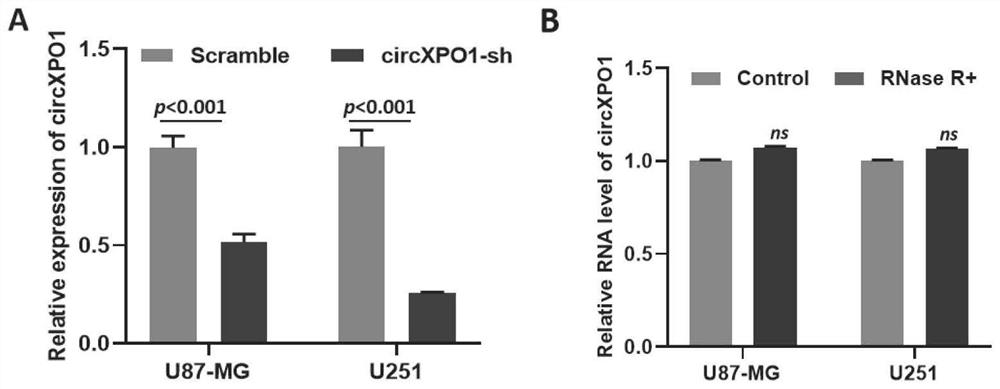
[0041] Example 3: Inhibition of circXPO1 expression using shRNA in U87-MG and U251 cells
[0042] 1. Experimental method
[0043] 1. Construction of scramble, circXPO1-sh plasmid (GFP co-expression): the most commonly used interference plasmid construction method is used, through endonuclease (Biolabs), enzyme digestion shRNA sequence and P GreenPuro vector (Sangon), using T4 Ligase (Biyuntian) ligates to form a plasmid.
[0044] The method for obtaining the virus liquid: (1) Take two 1.5mL centrifuge tubes, add 30 μL DMEM and 30 μL Fectin (ThermoFisher). (2) Take another 2 1.5mL centrifuge tubes, add 60μg DMEM, 0.75μg pspax2 packaging plasmid, 0.25μg pmd2.G packaging plasmid, add 1μg scramble, circXPO1-sh interference plasmid respectively, and then mix with the solution in step (1) centrifuge tube, After standing for 20 minutes, the cells were added to the 293T cell liquid cultured in DMEM medium containing 10% fetal bovine serum, respectively. After 24 and 48 hours, the su...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com