Construction method of multigene carrier and its application
A construction method and multi-gene technology, applied in the direction of using vectors to introduce foreign genetic material, DNA preparation, recombinant DNA technology, etc., can solve the problem that there is no effective method for gene assembly
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0102] Embodiment 1: accept the construction of carrier
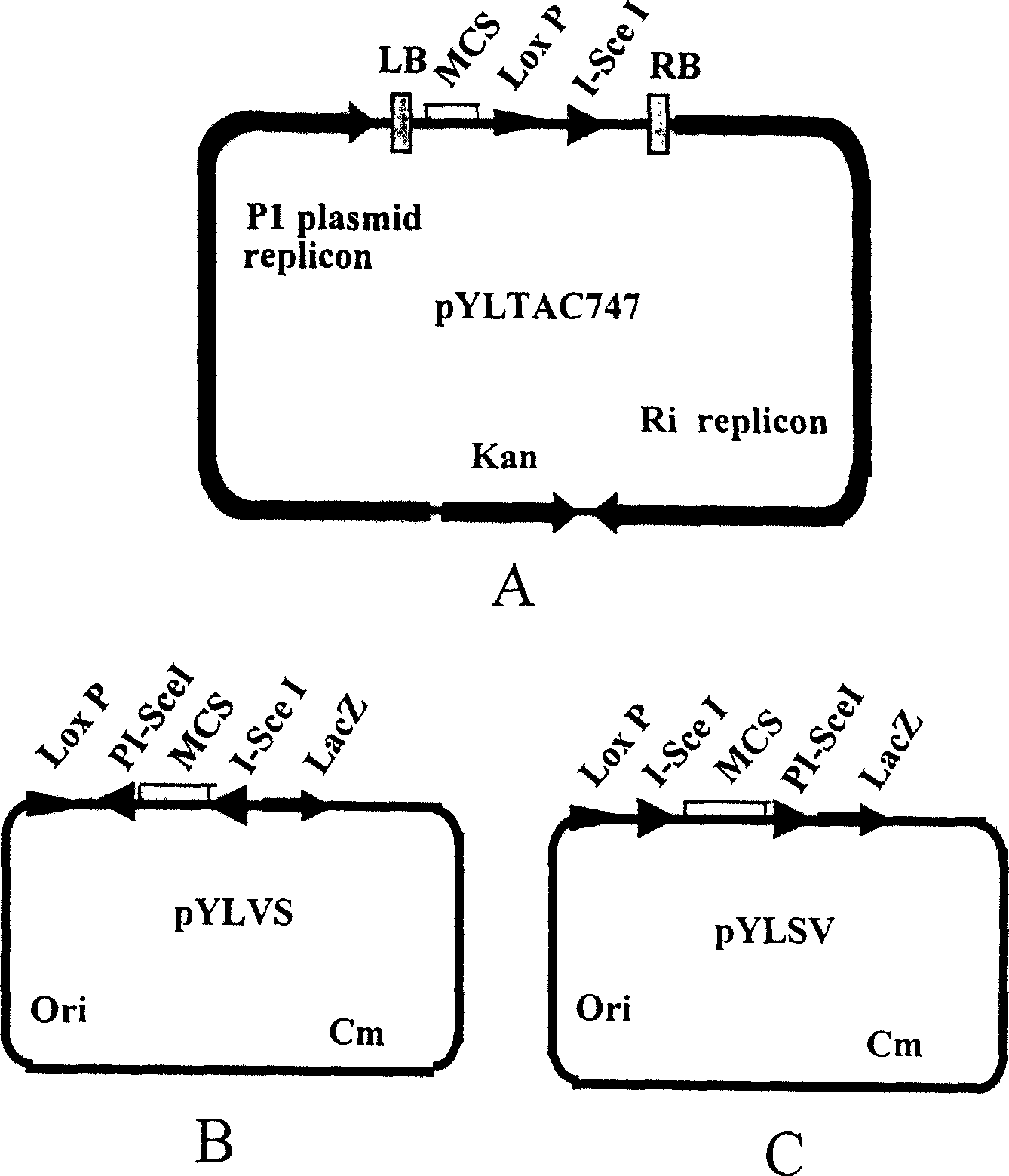
[0103] Such as image 3 As shown, according to the sequence of transformable artificial chromosome vector pYLTAC7 (Liu et al., 1999), primers P1 and P2 containing XbaI and BamHI sites were synthesized (see image 3 15690bp vector backbone fragment was amplified by PCR method, the cohesive end was cut out with XbaI and BamHI, and then ligated with the synthetic double-stranded DNA fragment MCS-LoxP-I-SceI (sequence 1 in the sequence listing) to form Circular, transform Escherichia coli DH10B to obtain acceptor vector pYLTAC747.
Embodiment 2
[0104] Embodiment 2: Construction of supply vector I and supply vector II
[0105] Such as Figure 4 As indicated, primers P3 and P4 were synthesized according to the sequence of the chloramphenicol resistance gene (see Figure 4Description), from the plasmid pCAMBIA1200 (CAMBIA company) PCR amplified 826bp chloramphenicol resistance gene Cm. Primers P5 and P6 were synthesized according to the sequence of plasmid pBluescript SK (ClonTech Company), and a 1660bp Ori-MCS-LacZ fragment was amplified from plasmid pBluescript SK by PCR. The two fragments were connected into a circle, transformed into Escherichia coli DH10B, and an intermediate product plasmid pYL was obtained. Use the restriction endonuclease BssHII to excise the MCS in the plasmid pYL, connect it with the synthetic double-stranded DNA fragment LoxP-PI-SceI-MCS-I-SceI (sequence 2 in the sequence listing) into a circle, and transform Escherichia coli DH10B , to obtain a donor vector I plasmid, named pYLVS. A rest...
Embodiment 3
[0106] Embodiment 3: the assembly of multigene transformation vector
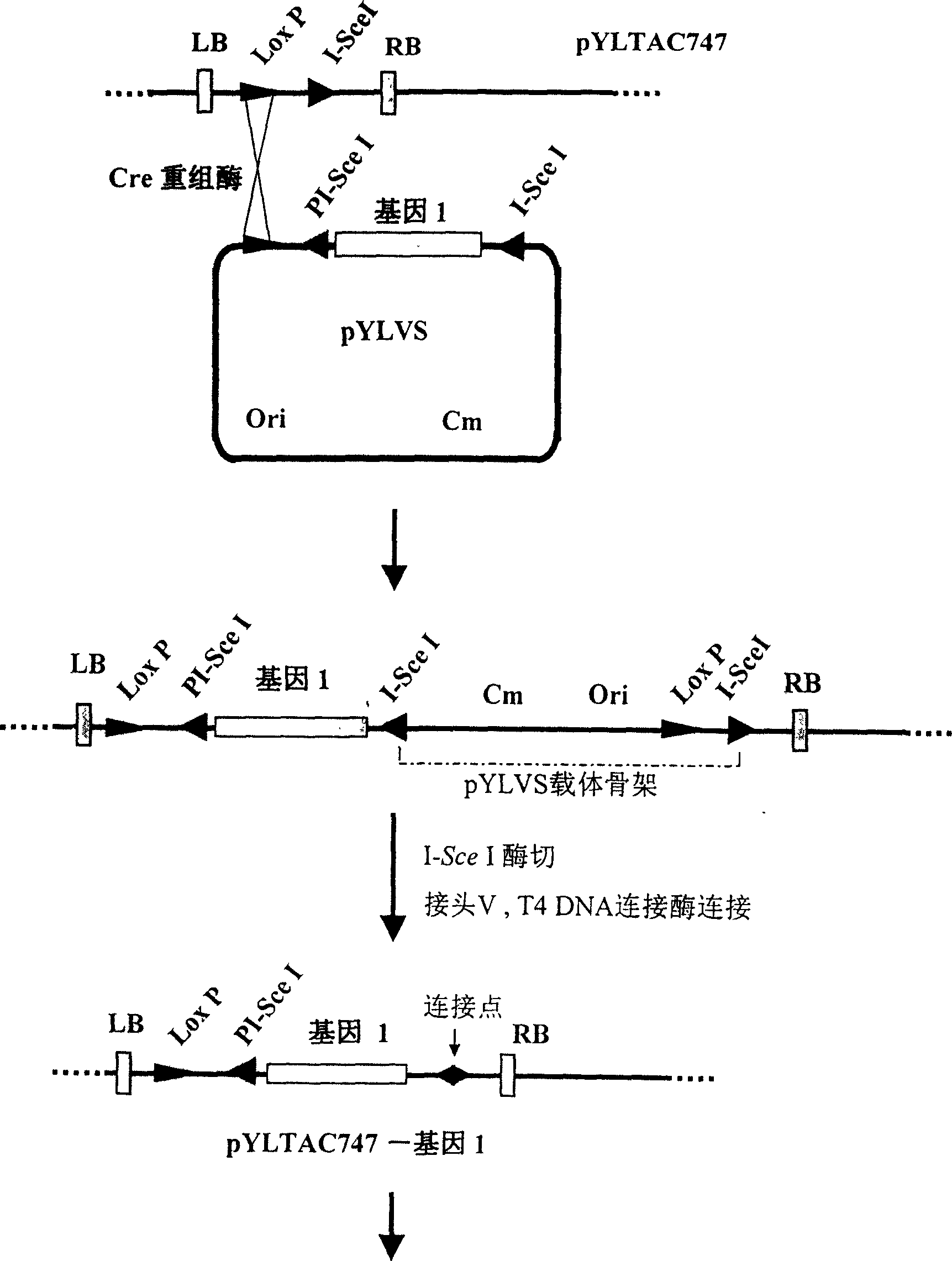
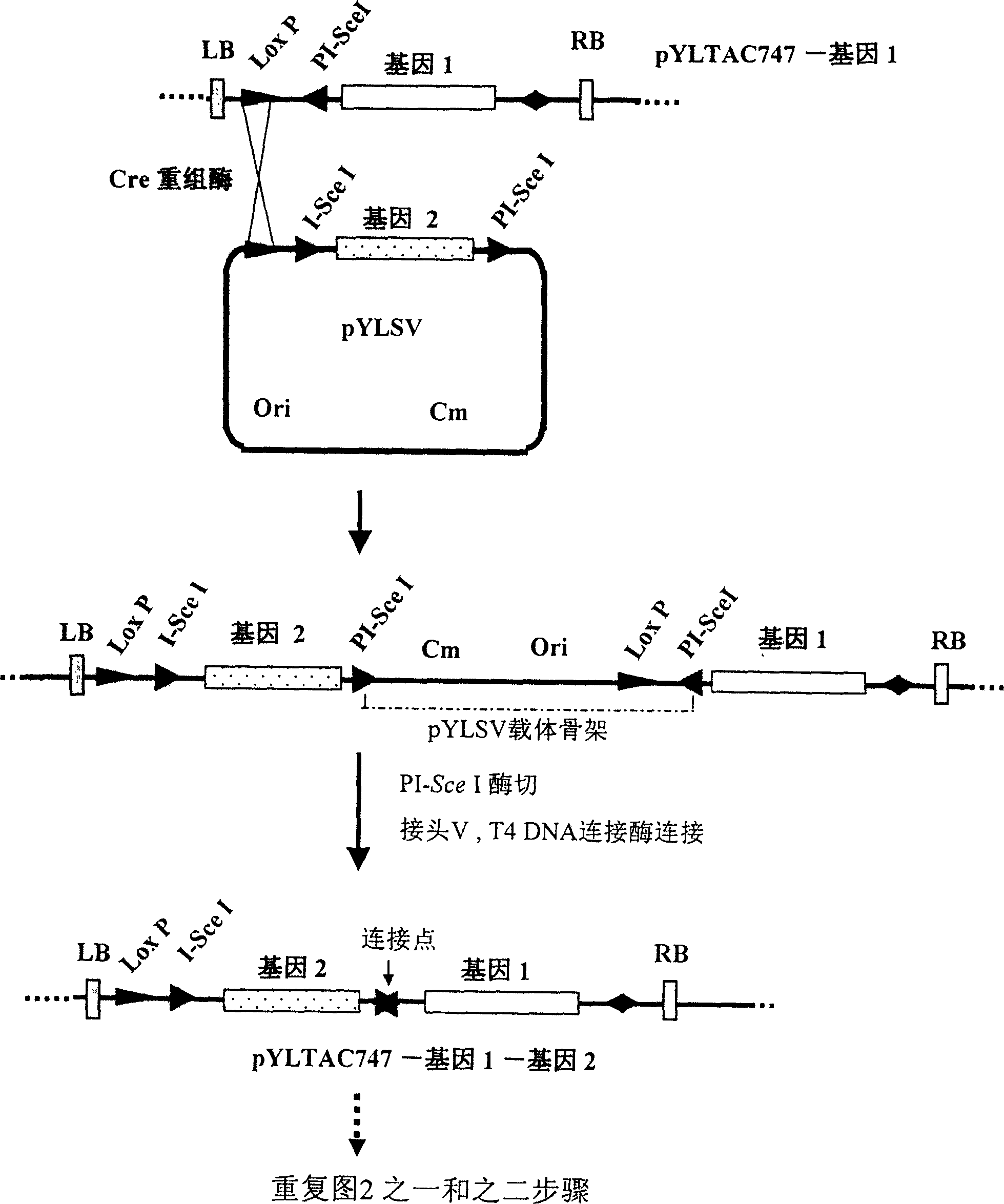
[0107] (1) Multiple cloning sites are set on the accepting vector pYLTAC747, and a few genes can be directly cloned into the vector by conventional molecular cloning methods. In this example, firstly, the hygrozyme gene HPT was directly subcloned into the NotI site in the cloning site of pYLTAC747. The resulting vector pYLTAC747HPT was as Figure 5 Swimming lane 2 is shown.
[0108] (2) Subcloning the MAR sequence (1.2 kb) into the donor vector I plasmid pYLVS to generate pYLVS-MAR. pYLVS-MAR and pYLTAC747-HPT were co-transformed into E. coli NS3529 competent cells containing Cre recombinase gene, so that the two plasmids were recombined and integrated in E. coli cells. The integrated plasmid was selected with kanamycin and chloramphenicol double antibodies, and then transformed into Escherichia coli DH10B without Cre enzyme gene. Use I-SceI to excise the pYLVS backbone, and use T4DNA ligase to ligate t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com