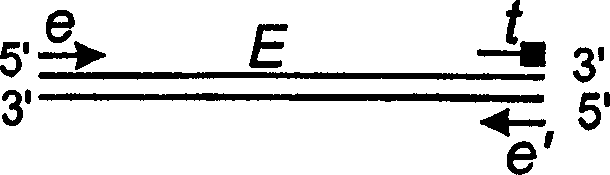Multi-local genomic analysis by method of improved cycle sequencing
A sequencing primer and nucleic acid sequencing technology, applied in the field of nucleic acid analysis, can solve problems such as diseases and cell collapse
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
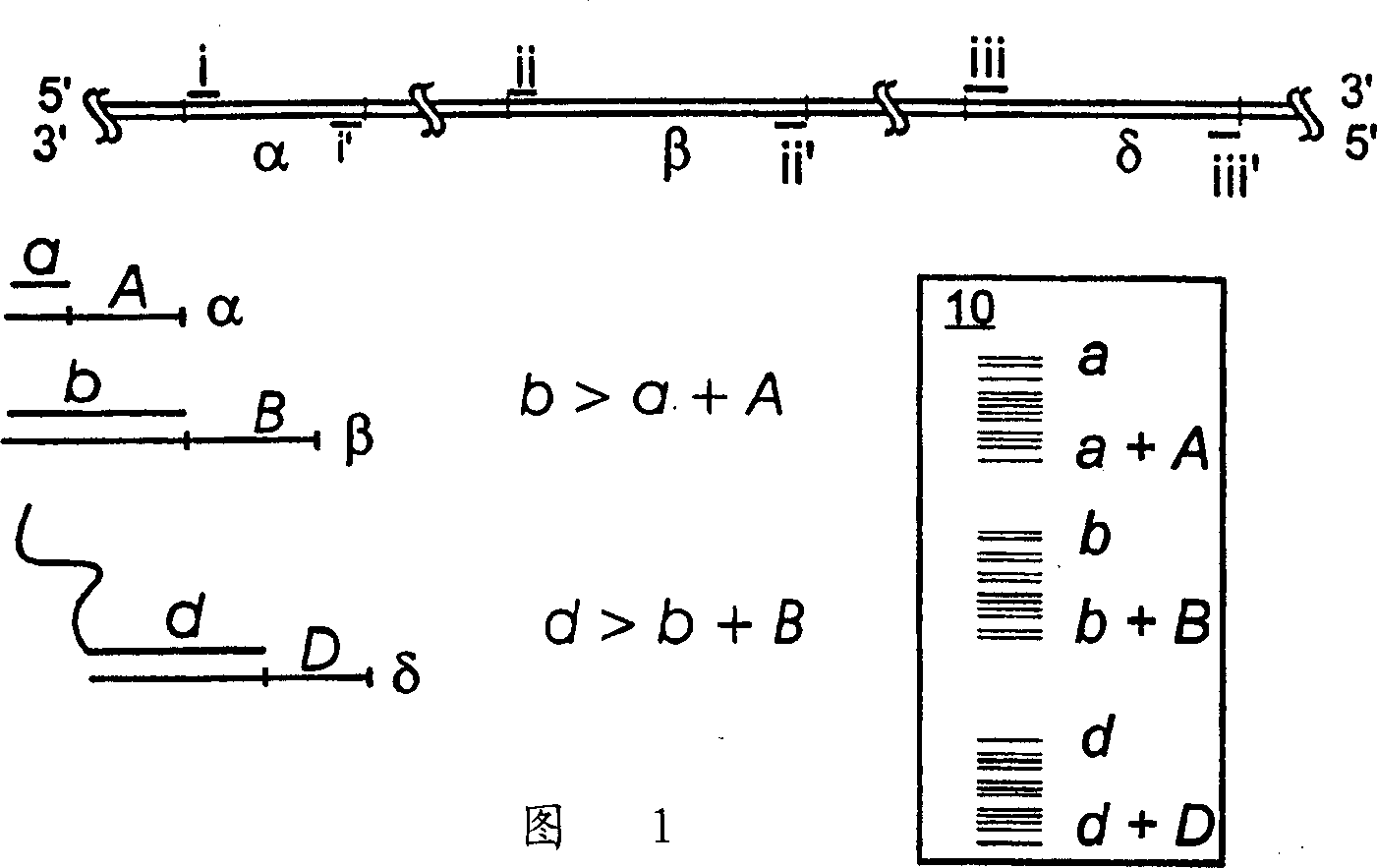
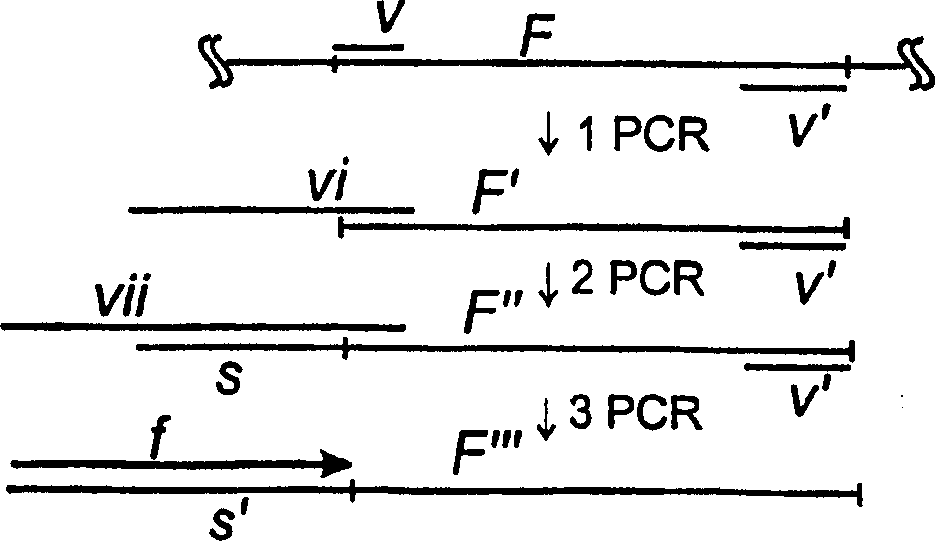
Image
Examples
Embodiment 1
[0057] Disclosed in this example are aspects of the invention that can be used to detect bovine viral diarrhea virus (BVDV). The method of this example can be adapted to detect and identify nucleic acids from other organisms, such as other RNA viruses.
[0058] The sequence analysis of this example employs a two-dye system. The system utilizes two lasers to detect the labeled products of its sequencing reactions. One laser was excited at a wavelength of 700nm and the other at a wavelength of 800nm. The primers of the sequencing reaction are labeled with two different near-infrared fluorescent dyes (such as dyes purchased from LI-COR, Inc. of Lincoln, Nebraska, U.S.A.), and one dye can respond to the irradiation of 700nm laser, which is recorded as IRD700 , another dye can react to 800nm laser, denoted as IRD800. Using this system, two different DNA fragments of the same molecular size produced by different sequencing reactions but each labeled with a different dye can be ...
Embodiment approach
[0087] cDNA can be prepared from isolated RNA according to methods known in the art. One implementation is as follows:
[0088] a. Mix the following reagents in a 1.5ml centrifuge tube:
[0089] Total RNA 1-2μl;
[0090] Downstream primer (10pM / ml) 2μl;
[0091] Water 8μl
[0092] b. Heat the mixture to 70°C for 10 minutes;
[0093] c. Add the following reagents to the heated mixture:
[0094] 4 μl of 5× first-strand synthesis buffer;
[0095] dNTP (10mM) 1 μl; 0.1M DTT 2 μl;
[0096] d. Heat the mixture to 42°C for 2 minutes;
[0097] e. Add 1 μl SUPERSCRIFT II (a reverse transcriptase available from LifeTechnologies, Inc., Gaithersburg, Maryland, U.S.A.) to the above mixture;
[0098] f. 42 ℃ heat preservation for 50 minutes;
[0099] g. Heat to 70°C and hold for 10 minutes to terminate the reaction.
[0100] 3. Amplification of Specific Segments
[0101] Amplification of cDNA may be performed by any suitable method known to those skilled in the art. In the imple...
Embodiment 2
[0123] This example provides the analysis of four segments of the BVDV genome using primers of different molecular weights and different fluorescent labels. This embodiment is to use the dual-dye sequencing system described in Example 1 to resolve the co-migration sequencing reaction products produced by primer sets 1 and 2, and to distinguish the co-migration sequencing reaction products produced by primer sets 3 and 4, while the primer set The extension products of 1 and 2 were distinguished from the extension products of primer sets 3 and 4 by molecular weight. The primers used in this embodiment are as follows:
[0124] Primer set 1
[0125] [IRD700 labeled] 5'GTA GGT AGA GTG AAA CCC GG
[0126] (Hybridizes with the first strand of the BVDV genome at position 6941-6961)
[0127] [Unlabeled] 5' CGG GAC CTG GAC TTC ATA GC
[0128] (can hybridize with the complementary strand of the BVDV genome at position 7255-7245)
[0129] Primer set 1 can amplify a segment of 314 bas...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com