DNA synthesis sequencing method and system
A technology of synthesis sequencing and control system, applied in the field of genetic engineering, can solve the problems of slow extension reaction, slow speed, accumulation of error information, etc., and achieve the effect of clean reaction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] Embodiment 1: Synthesis of water-soluble bifunctional linking unit:
[0086] Connecting unit 1 is directly purchased as shown in formula I.
[0087]
[0088] The connection unit 2 is shown in formula II,
[0089]
[0090] Its synthetic route is as follows:
[0091]
[0092] Concrete synthetic steps are:
[0093] 1. Dicyclohexylcarbodiimide (DCC) (80mg; 0.4mmol) and N, N-dimethylaminopyridine (3mg) were added to a solution of ethyl acetate (3mL) dissolved in tribromopropionic acid (compound 1) (43mg, 0.28mmol), at 20 Stir for 5min under the protection of argon at ℃, then add N-hydroxysuccinimide (compound 2) (72mg; 0.6mmol) and continue stirring for 1h, filter the reaction solution, wash with ethyl acetate, concentrate the filtrate, and purify it with a silica gel column to obtain the product (Compound 3) 3-bromo-2,5-oxo-1-pyrrolidinyl ester;
[0094] 2. Dibromoethanol (compound 4) (187.5 mg, 1 mmol) and KOH (33 mg, 0.6 mg) were added to 25 mL of DMSO solven...
Embodiment 2
[0122] Embodiment 2: the synthesis of connecting unit and reversible terminator
[0123] connection unit The synthetic route of is as follows:
[0124]
[0125] The specific synthesis steps are as follows:
[0126] 1) Weigh ethylene glycol (6.7ml, 120mmol) and acetic acid (2.4g, 40mmol) into a 100mL single-necked flask and stir, add 0.112mL of concentrated sulfuric acid dropwise to the reaction solution, and stir at 25°C for 24h. Then add 17mL of saturated sodium bicarbonate solution and stir overnight, then add 12mL of water to the reaction mixture, extract with dichloromethane (50mL*8), combine the organic layers, dry with anhydrous sodium sulfate, spin off the solvent, and the residue Use 30:1CH 2 Cl 2 / MeOH was used as the eluent for column chromatography to obtain compound 1 (2.52 g), with a yield of 61%. 1 H NMR (400MHz, CDCl 3 ): δppm4.20(t, 2H, J=4.8Hz), 3.82(t, 2H, J=4.8Hz), 2.09(s, 3H), 1.93(s, 1H).
[0127] 2) Weigh 2-bromoethanol (9.96g, 80mmol) and sodium...
Embodiment 3
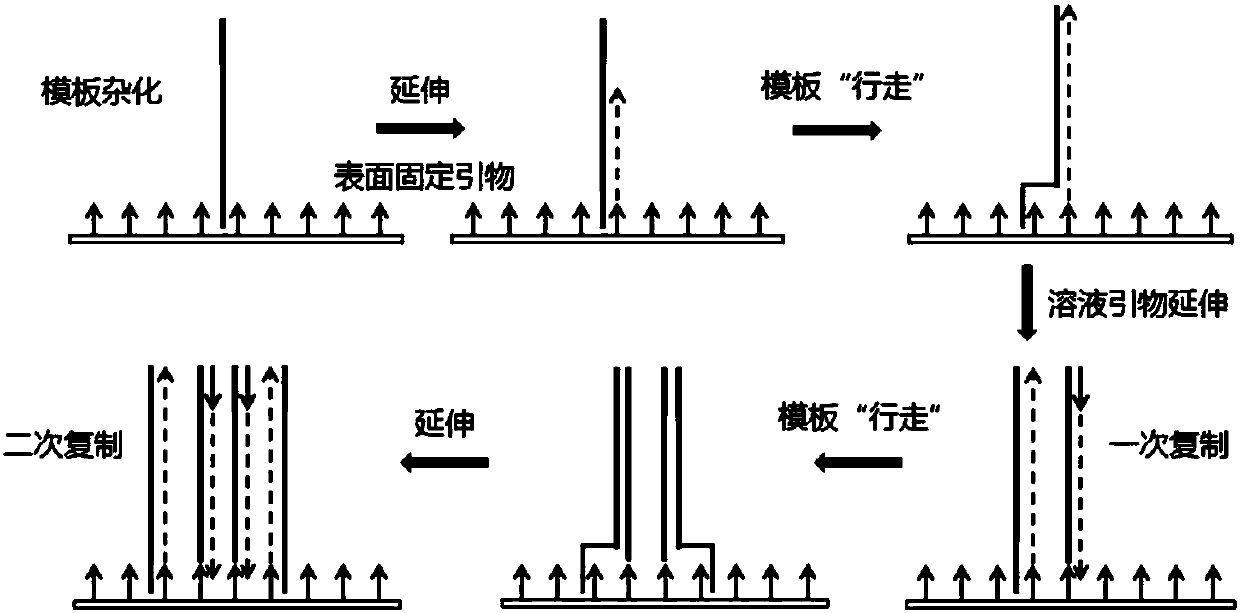
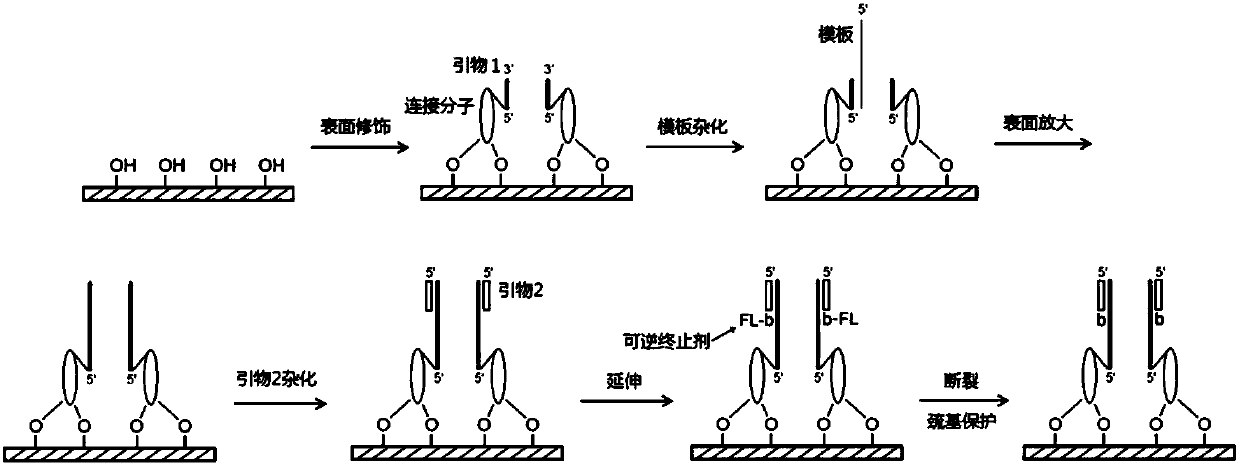
[0135] Example 3: Isothermal Amplification Surface Amplification
[0136] (1) Activate the surface of the slide to make it hydroxylated
[0137] The slides (4×4mm) were washed three times with ethanol and water respectively, dried and placed in hydrogen peroxide (H 2 o 2 , 30%) and concentrated sulfuric acid (H 2 SO 4 , 98% of the mixture (V (H 2 o 2 ):V(H 2 SO 4 )=7:3), heat at 80-90°C for 1 hour, take it out, rinse with plenty of water, and blow dry.
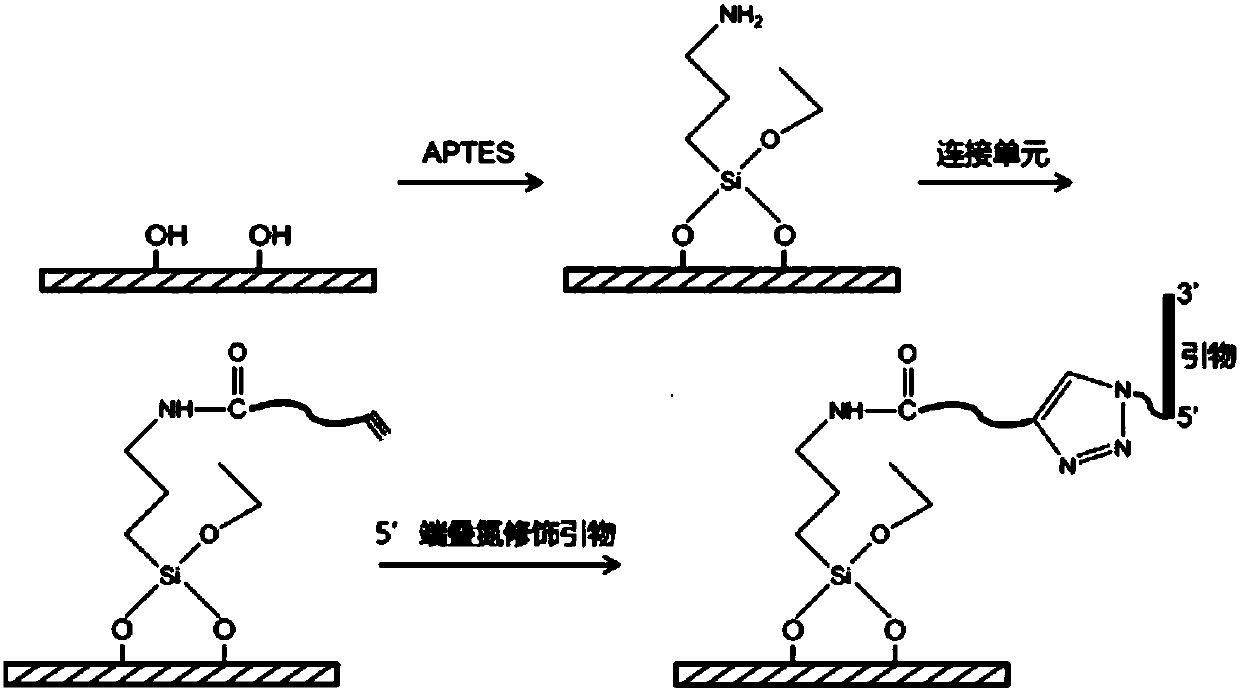
[0138] (2) Surface modification of glass slides
[0139] Place the above treated slides in absolute ethanol; add aminopropyltriethoxysilane (APTES) Make the mass ratio of APTES to absolute ethanol in the system 1:49, raise the temperature to 60°C, heat for 2h, and then cool to room temperature; APTES is connected to the glass surface through a silicon-oxygen-silicon bond, and then rinsed with ethanol and pure water respectively to obtain Glass slides with amino groups on the surface; immerse the glass slides with am...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com