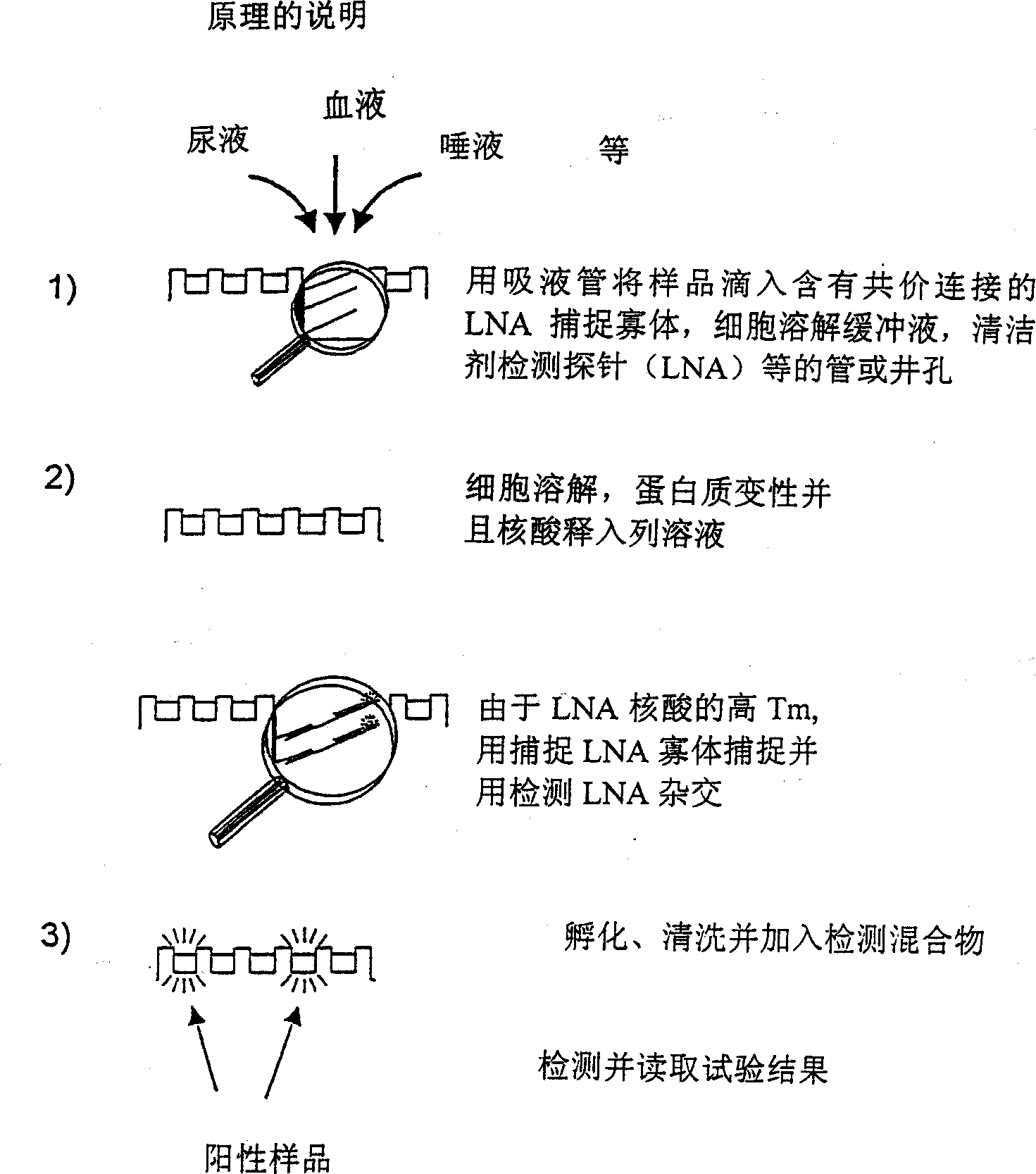
One step sample preparation and detection of nucleic acids in complex biological samples
A sample and nucleic acid technology, applied in the 0-24 field, can solve problems such as labor consumption, time-consuming, skin corrosion, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
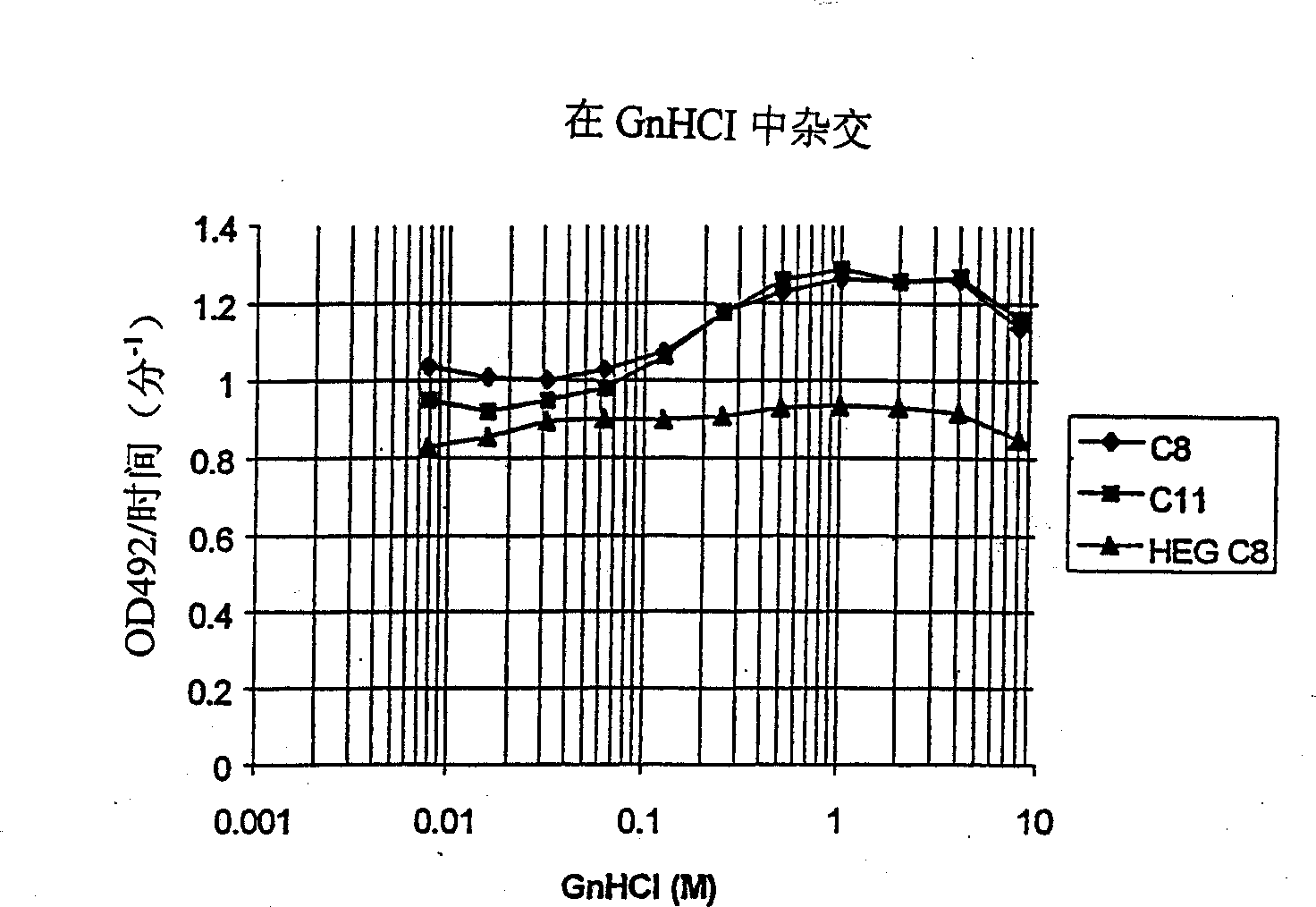
[0110] Example 1 GnHCl enables and promotes LNA hybridization in phosphate buffer
[0111] To investigate the role of strong chaotropic agents such as guanidine hydrochloride (GnHCl) in hybridization, the following experiments were performed.
[0112] LNA-modified oligomers with 5' anthraquinones (see Table 1-1) are covalently immobilized on the wells of a microtiter plate by UV (ultraviolet) irradiation, and are analyzed by hybridization with complementary target DNA oligomers used as capture probes. The hybridized material is detected by a 5' biotinylated DNA detection probe contained in the hybridization mixture.
[0113] name
EQ number
SEQ ID
NO
sequence
feature
C8
EQ-3133
4
5’-AQ-tac atg tta tgc ttt
GAC met C met GT GTg-3'
5'Anthraquinone modification
LNA
C11
EQ-3131
5
5’-AQ-tac atg tta tgc ttt
AAG AC met C met GTG
TG...
Embodiment 2
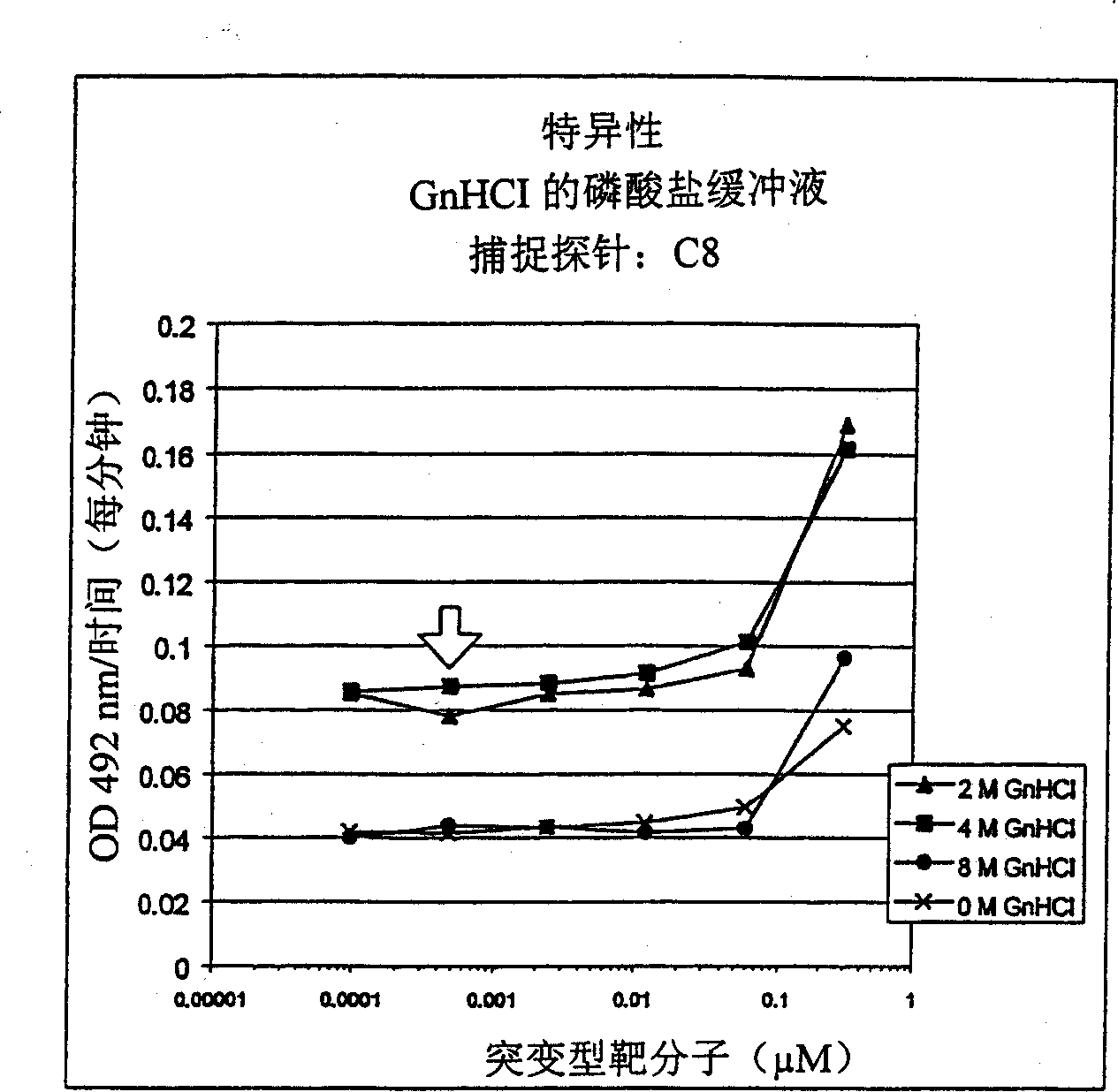
[0122] Hybridization specificity (competition experiment) in embodiment 2GnHCl
[0123] To investigate whether hybridization in a buffer containing a strong chaotropic agent such as guanidine hydrochloride (GnHCl) can exhibit sufficiently high stringency that single bases can be isolated, the following experiments were performed.
[0124] name
EQ number
SEQ ID
NO
sequence
feature
C8
EQ-3133
4
5’-AQ-tac atg tta tgc ttt
GAC met C met GT GTg-3'
5' anthraquinone modified
LNA
T8
EQ-3134
9
5’-AQ-tac atg tta tgc ttt
GAC met T GT GTg-3'
5' anthraquinone modified
LNA
target molecule
EQ-3185
7
5’ttg aat tcc aag agc aca
cgg tct tca gtg aag ctg cag
ggc act tcc aa3'
wild type, intended
Meaning g / c pos.
9756 (50-mer)
target molecule
EQ-3187
10
5’ttg aat tcc a...
Embodiment 3
[0137] Embodiment 3GnSCN makes the hybridization in sodium citrate and phosphate buffer saline solution possible and promotes it to carry out
[0138] Standard lysis buffers for RNA preparation are usually based on sodium citrate buffer, eg Glisin (1974) Biochemistry 13, 2633 and Chirwin (1979) Biochemistry 18, 5294. The following experiments were used to compare the performance of hybridization in the presence of guanidinium thiocyanate (GnSCN) containing sodium citrate or phosphate based buffers.
[0139] name
EQ number
SEQ ID
NO
sequence
feature
C8
EQ-3133
4
5’-AQ-tac atg tta tgc ttt
GAC met C met GT GTg-3'
5' anthraquinone modified
LNA
T8
EQ-3134
9
5’-AQ-tac atg tta tgc ttt
GAC met T GT GTg-3'
5' anthraquinone modified
LNA
target molecule
EQ-3185
7
5’ttg aat tcc aag agc aca cgg
tct tca gtg aag ctg...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com