Plant-derived resistance gene
A plant and plant cell technology, applied in plant gene improvement, plant peptides, plant cells, etc., can solve problems such as difficult transfer
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
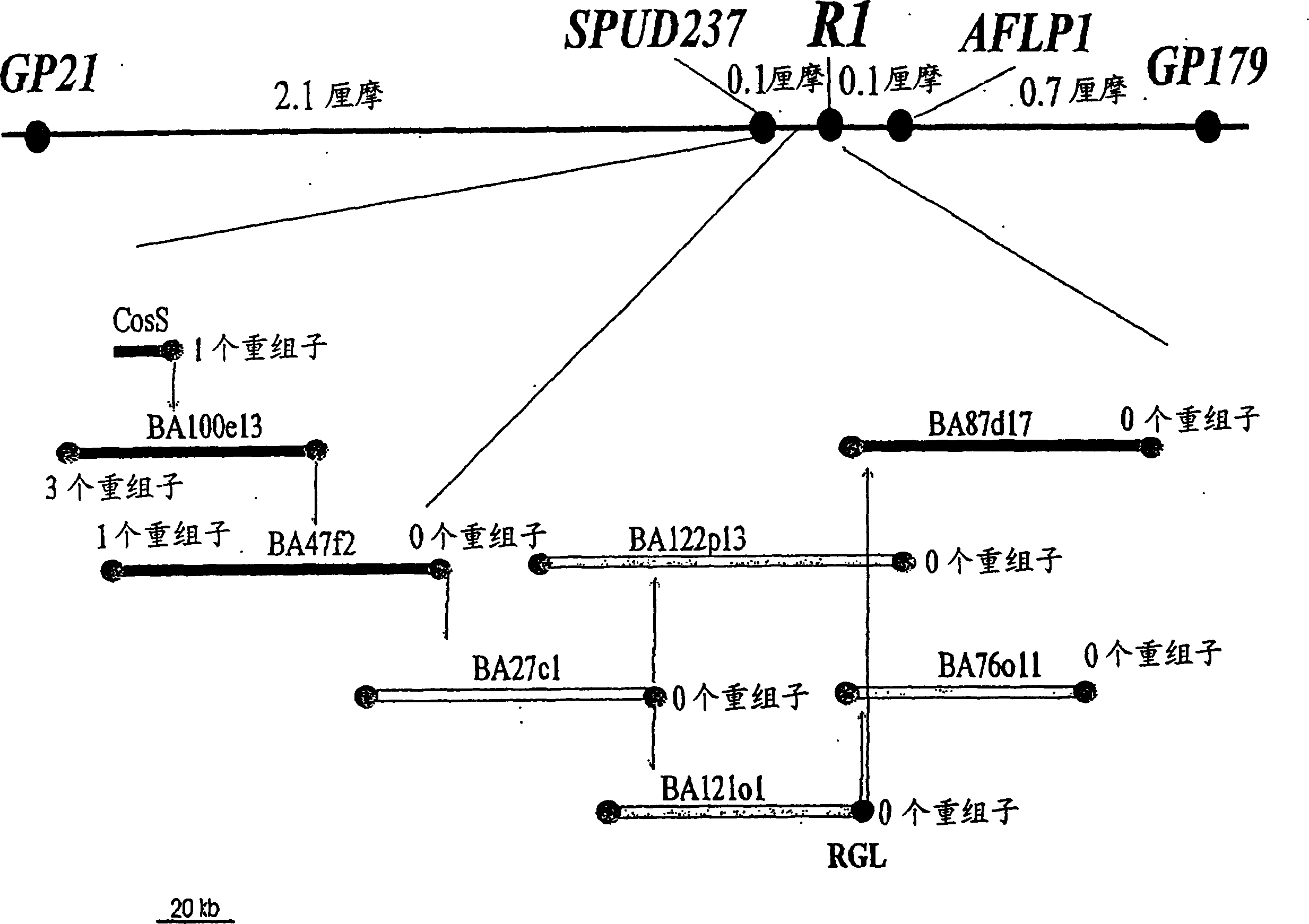
[0200] Example 1: High resolution genetic mapping of the R1 locus
[0201] To facilitate physical mapping of the R1 locus, 16 recombinants flanking R1 between the markers GP21 and GP179 (Leonards-Schippers et al., 1992) were selected from 588 plants, and their pairs carrying the corresponding Resistance of Phytophthora infestans to the avirulence factor Avr1. Together with 15 recombinants previously selected in the same interval (Meksem et al., 1995), there were a total of 31 recombinants in the GP21-GP179 interval in 1049 plants, corresponding to a recombination rate of 3.0% (3 cmM) . The recombination rates between GP21 and R1 and between R1 and Gp179 were 2.2% and 0.8%, respectively (Table 1).
[0202] Table 1. The number of recombinant individuals selected from the 1049 plants of the isolated F1 population in the GP21-R1, GP179-R1 and GP21-GP179 intervals
[0203] GP21-R1 GP179-R1 GP21-GP179
[0204] Number of recombinants 23 8 31
[0205] Recomb...
Embodiment 2
[0209] Example 2: Chromosomal Walking to the R1 Locus and Identification of R1 Candidate Genes
[0210] The marker SPUD237 was used as a probe for screening cosmid libraries. A positive clone CosS was identified ( figure 1 ). End-sequencing of the CosS insertion yielded a new marker that was separated from the R1 locus by a recombination event (0.1 centimorgan). Screening of the BAC library with this marker identified the BAC clone BA100e13. Three recombination events separate the distal end of BA100e13 from R1. BA47f2 was identified with the BA100e13 end close to R1. The end of BA47f2 distal from R1 overlaps with BA100e13 and is separated from R1 by a recombination event. Its proximal end co-segregates with R1, similar to the BAC end in all subsequent analyzes ( figure 1 the right part of the ). The BA27c1 clone was identified with the BA47f2 end co-segregating with R1. The BA27c1 ends that do not overlap with BA47f2 identified the BA122p13 and BA121o1 clones. The BA...
Embodiment 3
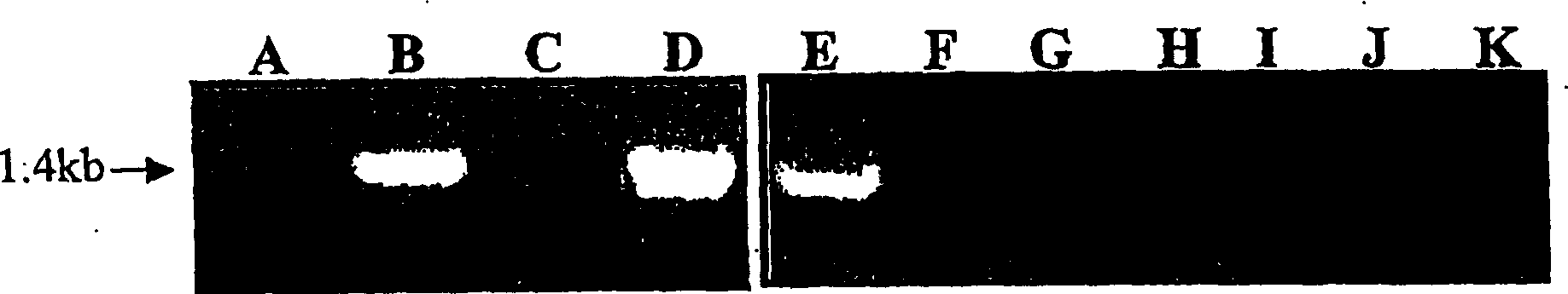
[0212] Example 3: R1 candidate cDNA clones
[0213] Using the complete inserts of BAC clones BA121o1 and BA76o11 as probes, six and eight cDNA clones were isolated, respectively, from cDNA libraries prepared from infested leaves of genotype P41(R1r1). Eight of these 14 cDNA clones were similar to known plant resistance genes. The highest similarity was obtained with the tomato gene Prf for resistance to Pseudomonas syringae (Salmeron et al., 1996). The sequences of these eight candidate cDNAs are approximately 80%-90% identical to each other. The genome sequence of the 2292 nucleotide long cDNA clone c76-2 is identical to the g10 clone (a subclone representing part of BA87d17), except for introns (see below). Sequence comparison with known resistance genes in databases showed not full length c76-2. Using RACE analysis, the cDNA was extended to the 5' end by 1943 nucleotides, and a full-length cDNA sequence of 4235 nucleotides was obtained. The full-length cDNA sequence cont...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com