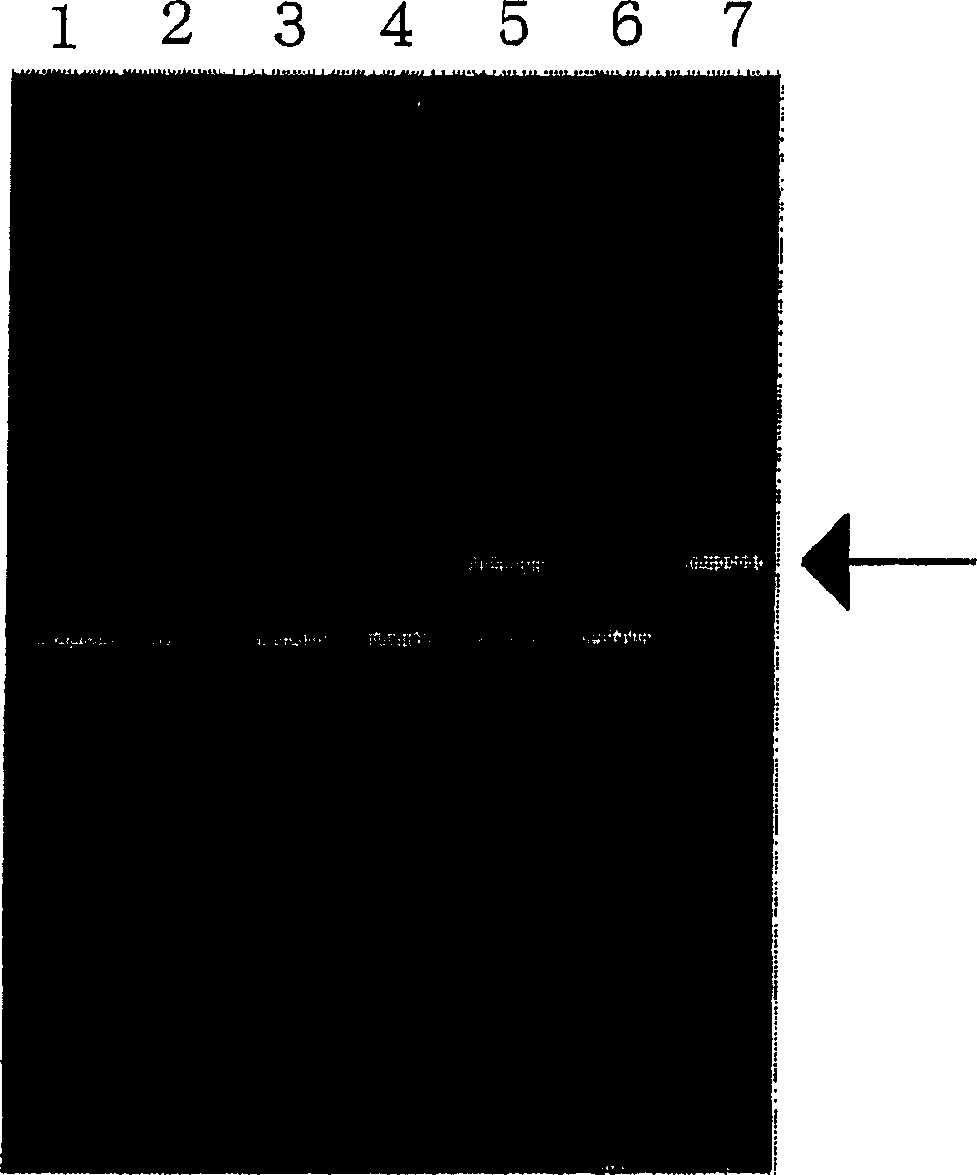Method of typing gene polymorphisms
A genetic polymorphism and gene technology, applied in the field of genetic polymorphism typing, can solve the problem that it cannot be used to detect trace target nucleic acids
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0176] An embodiment of the method for typing genetic polymorphisms in multiple target nucleic acids according to the present invention is exemplified below: a method for typing genetic polymorphisms, wherein multiple target nucleic acids are detected in parallel, the method comprising:
[0177] (1) template nucleic acid, deoxyribonucleoside triphosphate, DNA polymerase with strand displacement activity, at least two nucleotides and RNase H are mixed to prepare a reaction mixture, wherein
[0178] (a) the 3' terminus of said nucleotides is modified so that extension from that terminus does not occur under the action of a DNA polymerase; and
[0179] (b) the nucleotides each have a nucleotide sequence that anneals to a specific region containing the specific nucleotide in the plurality of target nucleic acids; and
[0180] (2) incubating the reaction mixture for a period of time sufficient to generate reaction products to extend nucleic acids each containing a region of arbitra...
Embodiment 1
[0241] (1) Preparation of genomic DNA
[0242]After obtaining informed consent from 7 healthy human donors, 7 samples of 10ml blood were collected. Genomic DNA was prepared from 100 µl of each of the above bloods using Dr. GenTLE (for blood) (Takara Bio.). The concentration of the obtained genomic DNA solution is as follows:
[0243] Test sample 1: 182ng / μl
[0244] Test sample 2: 150ng / μl
[0245] Test sample 3: 156ng / μl
[0246] Test sample 4: 204ng / μl
[0247] Test sample 5: 105ng / μl
[0248] Test sample 6: 172ng / μl
[0249] Test sample 7: 253ng / μl
[0250] (2) Synthesis of primers and probes
[0251] Design oligonucleotide primers that can be used for ICAN reaction detection and contain 3 RNA residues at the 3' end, and based on the nucleotide sequence of the human glutathione-S-transferase M1 (GSTM1) gene (GenBank No. X51451) synthesis. Specifically, oligonucleotide primers GS-F (SEQ ID NO: 6) and GS-R (SEQ ID NO: 7) were synthesized. Oligonucleotide GS-F is a ...
Embodiment 2
[0263] (1) Allele-specific detection of human CYP2C19(636)
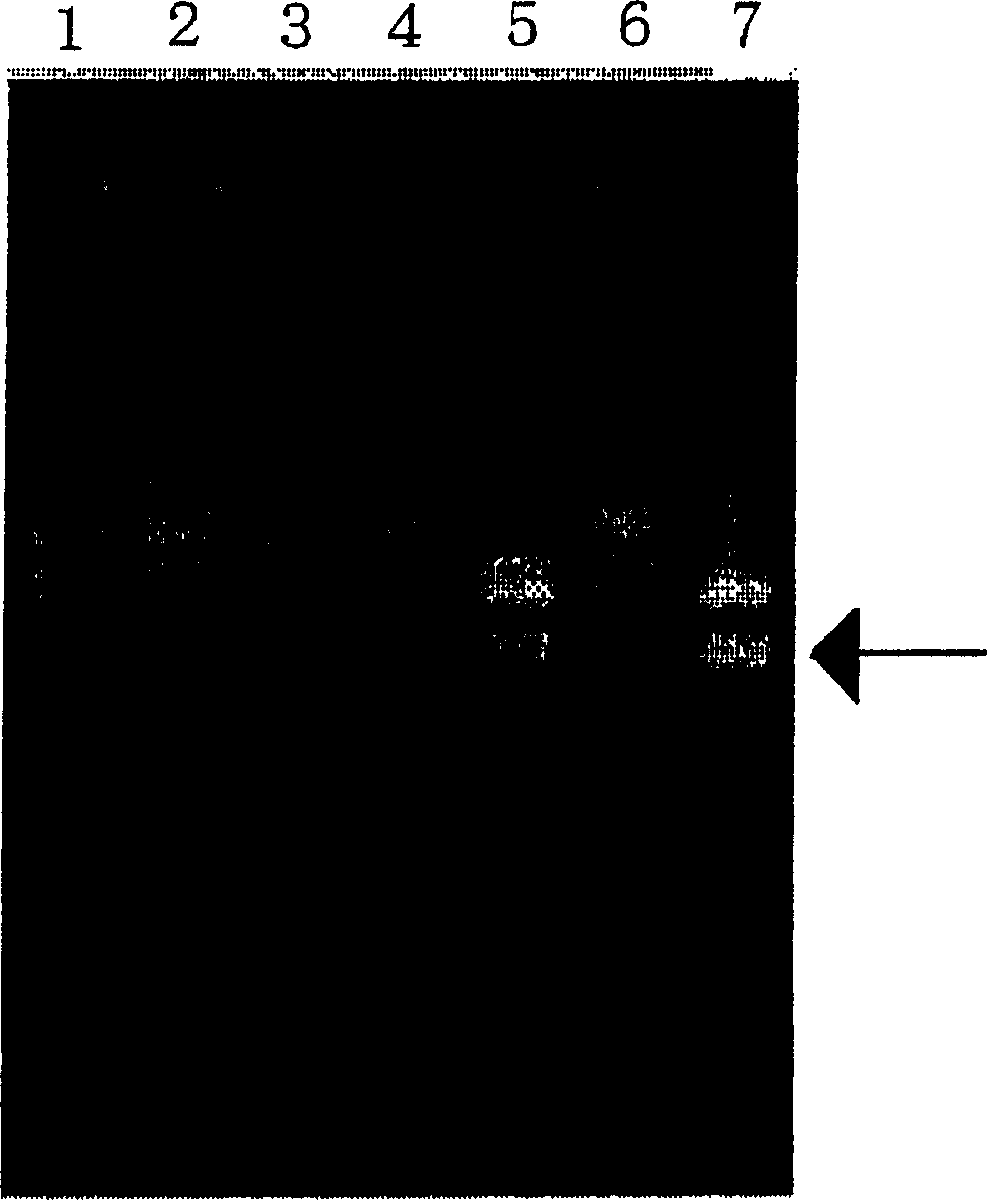
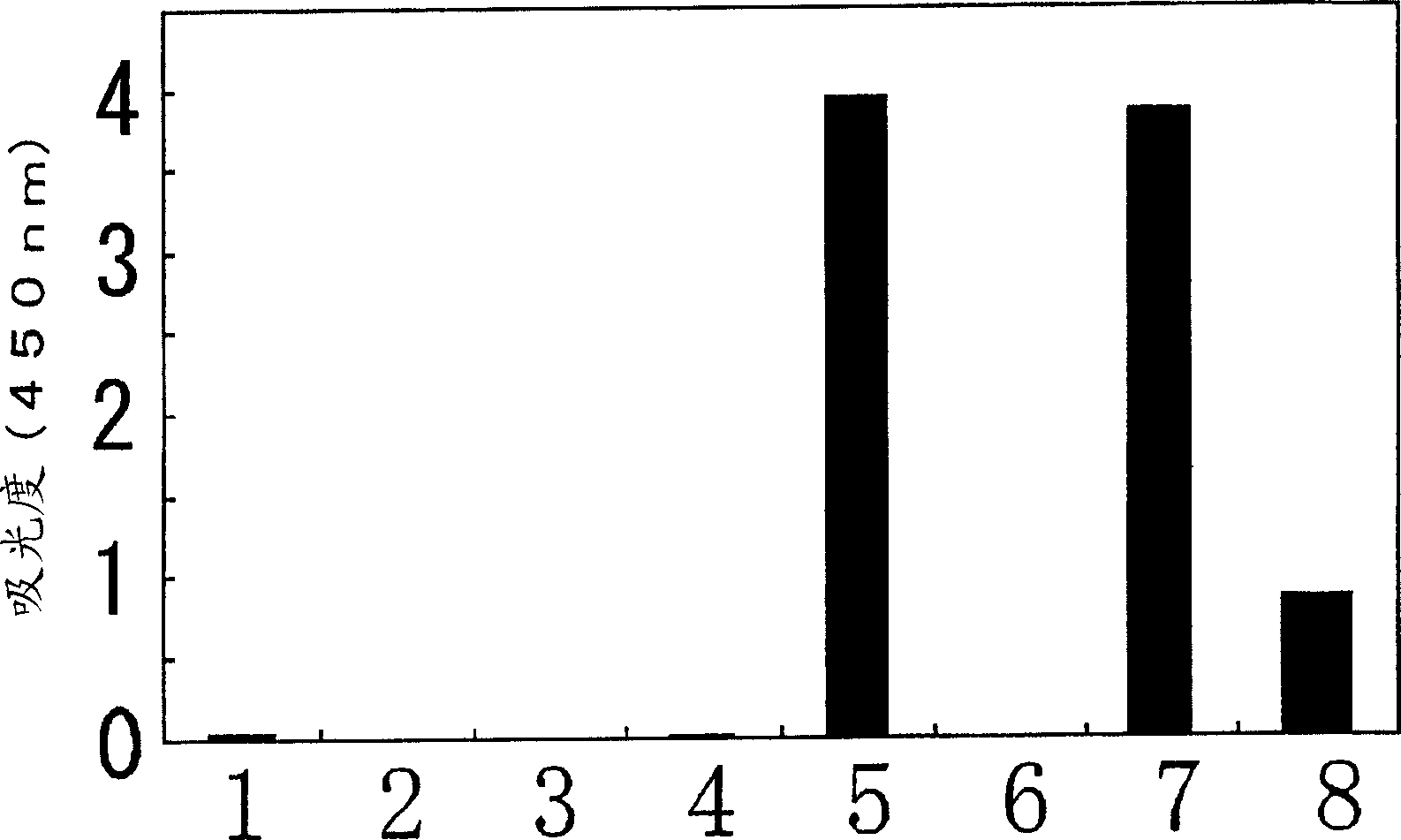
[0264] An assay for determining whether the allele at nucleotide 636 in human CYP2C19 is genetically homozygous or heterozygous (homozygous or heterozygous) was tested.
[0265] After obtaining informed consent, 200 μl whole blood was obtained from each healthy individual (samples 1-6) and processed using Dr.GenTLE TM (Takara Bio) Genomic DNA was prepared from the above whole blood. In a Thermal Cycler Personal (Takara Bio), a reaction mixture with a total volume of 5 μl containing 160 ng of prepared genomic DNA as a template, a sense primer for specific detection of 636G (SEQ ID NO : 14) or 636A (SEQ ID NO: 15) allele synthetic oligonucleotide and synthetic oligonucleotide (SEQ ID NO: 16) as an antisense primer: each 50 pmol, 1 μl 0.05% propylene imine aqueous solution , followed by annealing of the primers to the template at 53°C. 20 μl containing 0.625mM dNTP mixture, 40mM Hepes-KOH buffer (pH7.8), 125mM potass...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com