DNA molecular marking method for researching fish genetic relation
A technology of DNA molecules and genetic relationships, applied in the fields of botanical equipment and methods, biochemical equipment and methods, and applications, and can solve the problems of unrealistic sequencing of all species in the biological world.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
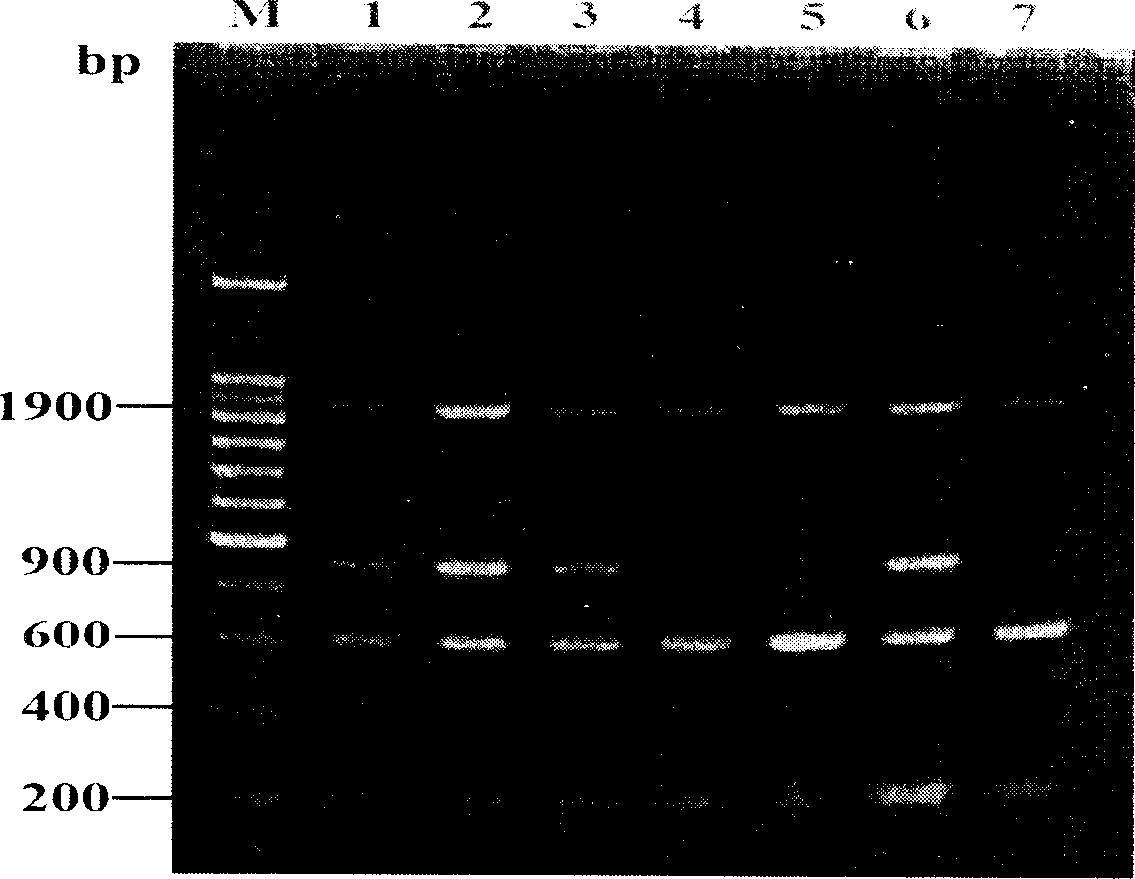
[0025] Now take the red crucian carp (Carassius carassius red var.) as an example to specifically introduce the present invention. The sex of red crucian carp was determined by vivisection, and the individual of red crucian carp was selectively matured. Blood was collected from the dorsal artery of red crucian carp with a disposable syringe, and blood DNA was extracted according to the UNIQ-10 Column Genomic DNA Extraction Kit from Shanghai Sangon. The concentration of the extracted DNA was detected by a UV spectrophotometer and stored at -20°C.
[0026] Referring to the conserved region sequence of Sox gene HMG in different species, unique degenerate primers were designed with Primer Premier 5.0 software and Jellyfish1.4 software, and synthesized by Shanghai Sangon Company. Its sequence is as follows: upstream primer: 5'TGAAGCGACCCATGAAC / TG3'; downstream primer: 5'AGGTCG(A / G)TACTT(A / G)TA(A / G)TT3'. Using the red crucian carp genomic DNA as a template, a PCR system was design...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com