Fluorescent quantitative PCR reagent kit for detecting epidermal growth factor receptor gene point mutation
A technology for epidermal growth factor and fluorescence quantification, which is applied in the field of molecular biology, can solve problems such as unsafety, hazards to operators and the environment, and complicated operation process, so as to reduce the probability of result deviation, reduce the possibility of pollution, and operate The effect of simple process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
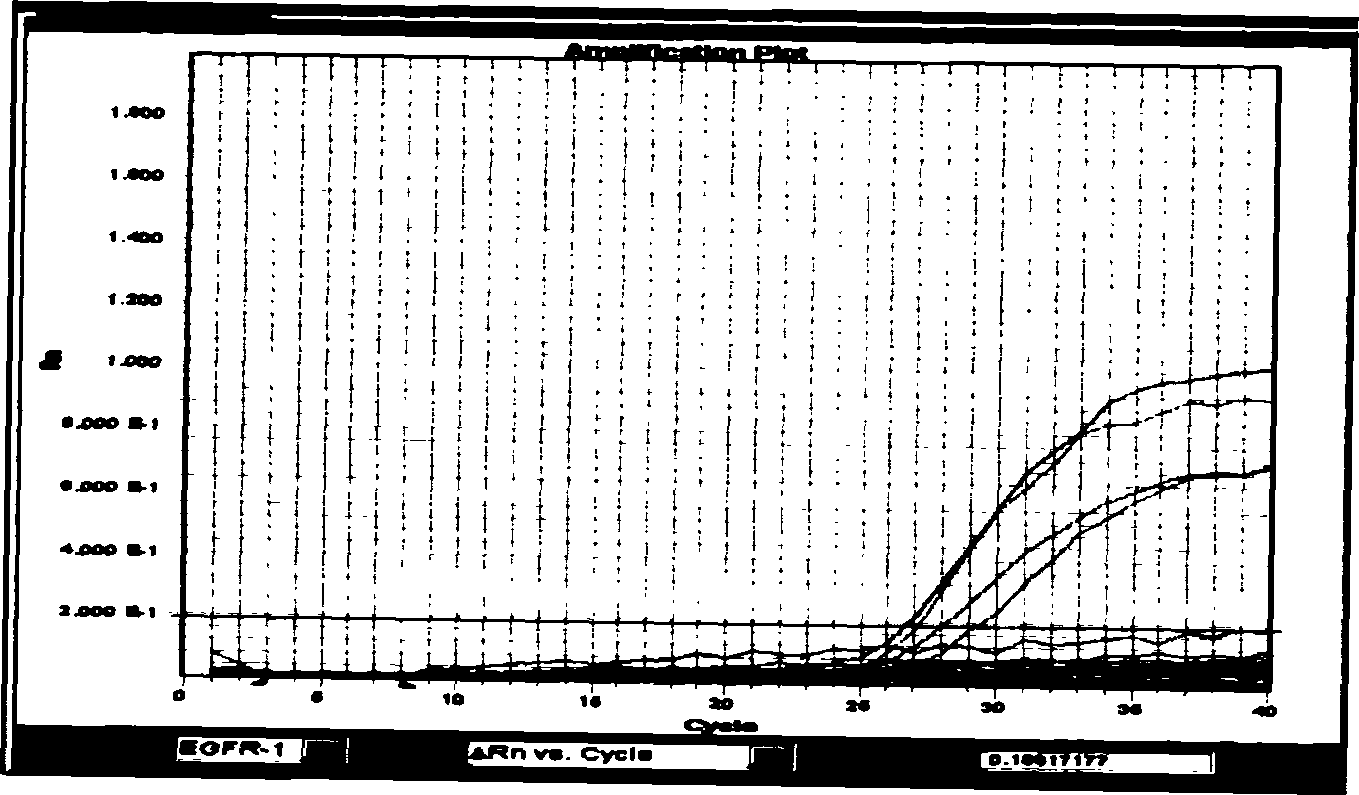
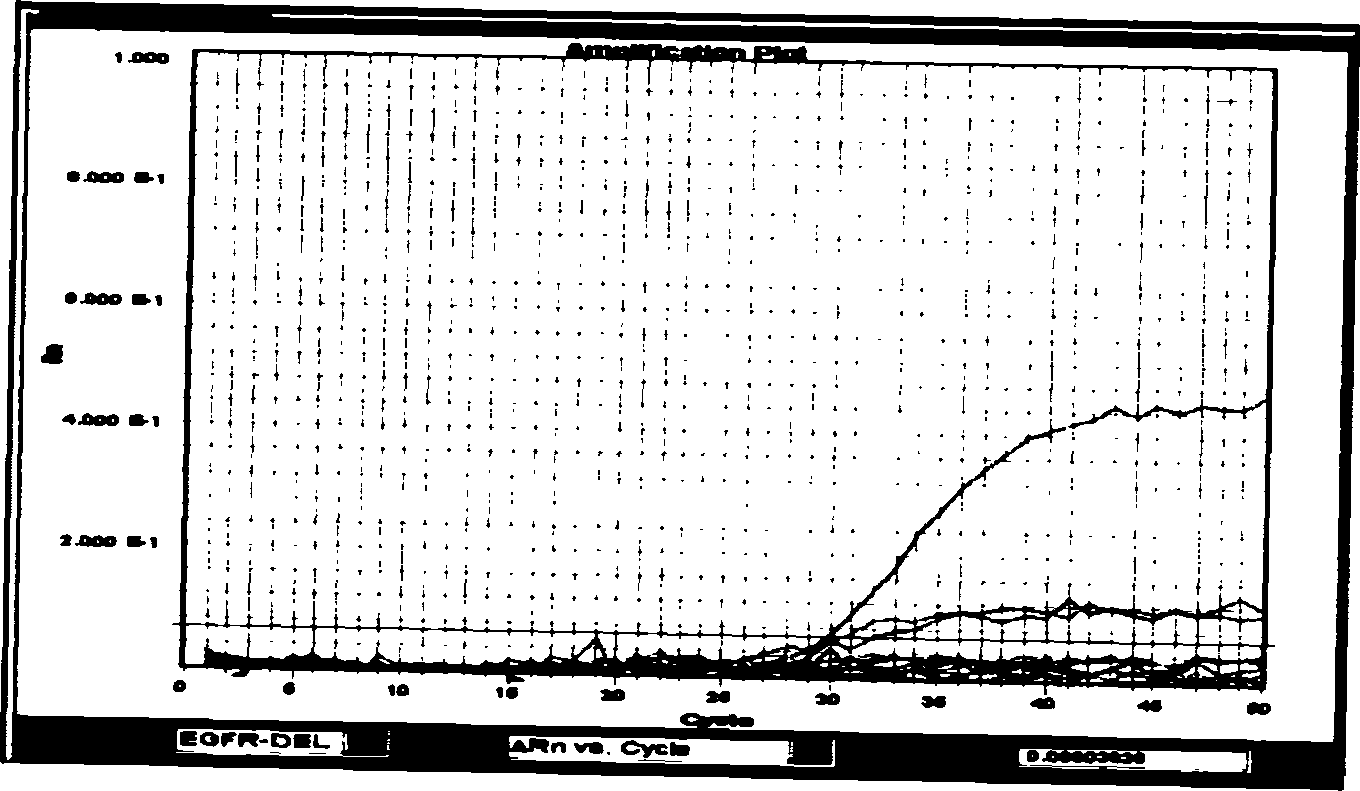
[0038] Example 1 Detection of two deletion mutations on exon 19 of the EGFR gene by fluorescent quantitative PCR (deletion of 15 bases at positions 2235-2249; deletion of 18 bases at positions 2240-2257):
[0039] Design a probe and a pair of common primers that can specifically detect 15 base deletions and 18 base deletions:
[0040] SEQ ID NO: A1 CTGGATCCCAGAAGGTGAGAAA
[0041] SEQ ID NO: A2AGCAGAAACTCACATCGAGGATTT
[0042] SEQ ID NO: A3 FCCGTCGCTATCAAAACATCTCCGAAP (probe for detecting deletion of 15 bases, F represents FAM group, P represents TAMRA group) SEQ ID NO: A46TCGCTATCAAGGAATCGAAAGCCAACP (probe for detecting deletion of 18 bases, 6 represents HEX group , P represents the TAMRA group).
[0043] Then optimize the reaction system for FQ-PCR detection: the reaction system is 15 μl, the forward and reverse primers are 0.15 μl (20 μM), the probe is 0.2 μl (20 μM), the template DNA is 2.5 μl (100-300ng / μl), 2* Taqman universal PCR Master Mix (purchased from Applied Bio...
Embodiment 2
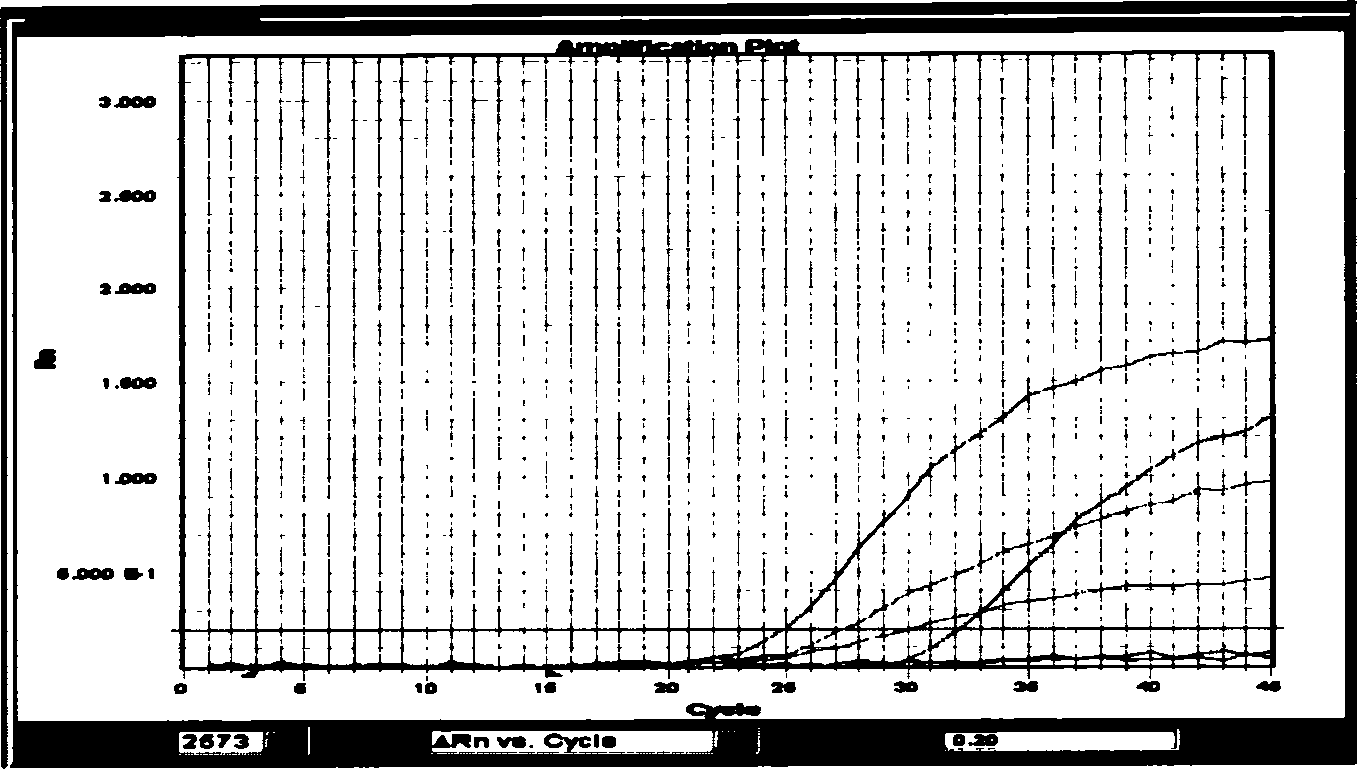
[0045] Example 2 Detection of two point mutations (base substitution mutation at position 2573 and base substitution mutation at position 2582) on exon 21 of the EGFR gene by fluorescent quantitative PCR: firstly, a design capable of specifically detecting the base at position 2573 One probe for base substitution mutation and 2582 base substitution mutation and one pair of common primers:
[0046] SEQ ID NO: B1 AACACCGCAGCATGTCAAGA
[0047] SEQ ID NO: B2 CCTTACTTTGCCTCCTTCTGCAT
[0048] SEQ ID NO: B3 FTTTGGCCCGCCCAAAATCTGTP (probe for detecting mutation at position 2573, (F represents FAM group, P represents TAMRA group). SEQ ID NO: B47CGCACCCAGCTGTTTGGCCP (probe for detecting mutation at position 2582, (7 represents TET group) group, P represents the TAMRA group).
[0049] Then optimize the reaction system for FQ-PCR detection: the reaction system is 15 μl, the forward and reverse primers are 0.15 μl (20 μM), each probe is 0.2 μl (20 μM), template DNA is 0.5 μl (100-300 ng / ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com