Polyspecific binding molecules and uses thereof
a technology of polyspecific binding molecules and binding molecules, which is applied in the direction of peptides, drug compositions, fused cells, etc., can solve the problems of inability to isolate sufficient quantities of ctls from patients, significant drawbacks in the approach, and additional complications
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of p-149 Single-Chain (sc) TCR
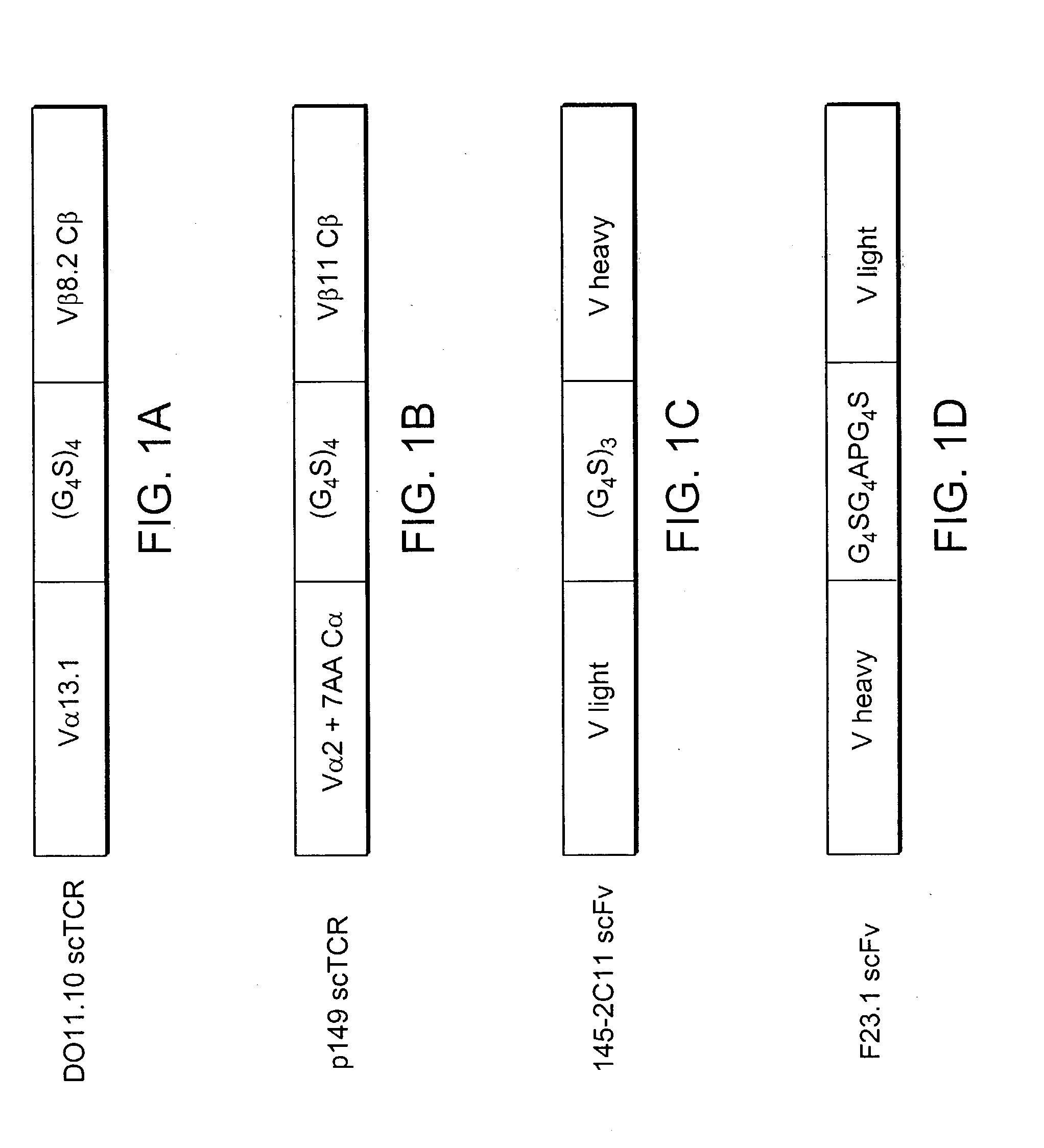
[0273] The T cell clone, p-149, recognizes a peptide fragment (STPPPGTRV, SEQ ID NO. 11) of the human wild-type tumor suppresser protein p53 restricted by HLA-A2.1. (See Theobald et al., PNAS, 1995) The T cell receptor gene was cloned into a three domain single-chain format previously shown to produce soluble TCR and functional receptor molecules (FIG. 1A).
[0274] In brief, mRNA was isolated from the T cell clone and cDNA was made using the Marathon cDNA Amplification Kit (Clontech). The cDNA was used as a template in polymerase chain reaction (PCR) with primers KC 171 and KC174 to produce a 5'SfiI3'SpeI V.alpha. chain fragment including the first seven amino acids of the C.alpha. chain N-terminus. The same cDNA was then used as a PCR template with primers KC172 and KC 176 to generate a 5'XhoI-3'XmaI V beta C beta chain fragment. The C beta chain was truncated just before the cysteine residue at amino acid 127 of the full-length C beta chain...
example 2
Purification and Characterization of the p-149 sc-TCR
[0285] The pending U.S. application Ser. No. 08 / 943,086 discloses a variety of methods for purifying sc-TCR proteins including these that comprise the D011.10 sc-TCR. These methods can be adapted to purify the p-149 fusion protein. For example, to purify the scTCR, an antibody with specificity for a conformational epitope on V.beta. 11.0 or V.alpha. 2.3 can be used along lines disclosed in the pending U.S. application. In particular, the p-149 scTCR can be purified on an immunoaffinity column using the following procedure.
[0286] Cell paste generated from a fermentor can be suspended in extraction buffer followed by mechanical lysing of cells by passage through a French press. The supernatant is clarified by centrifugation at 25,000.times.g and applied to a Q-sepharose column. The scTCR is collected in the flow-thru and then applied to a Protein-A-sepharose column cross-linked with mAb H57-95. This is a hamster mAb specific for an ...
example 3
Construction, Expression and Characterization of the DO11.10 scTCR
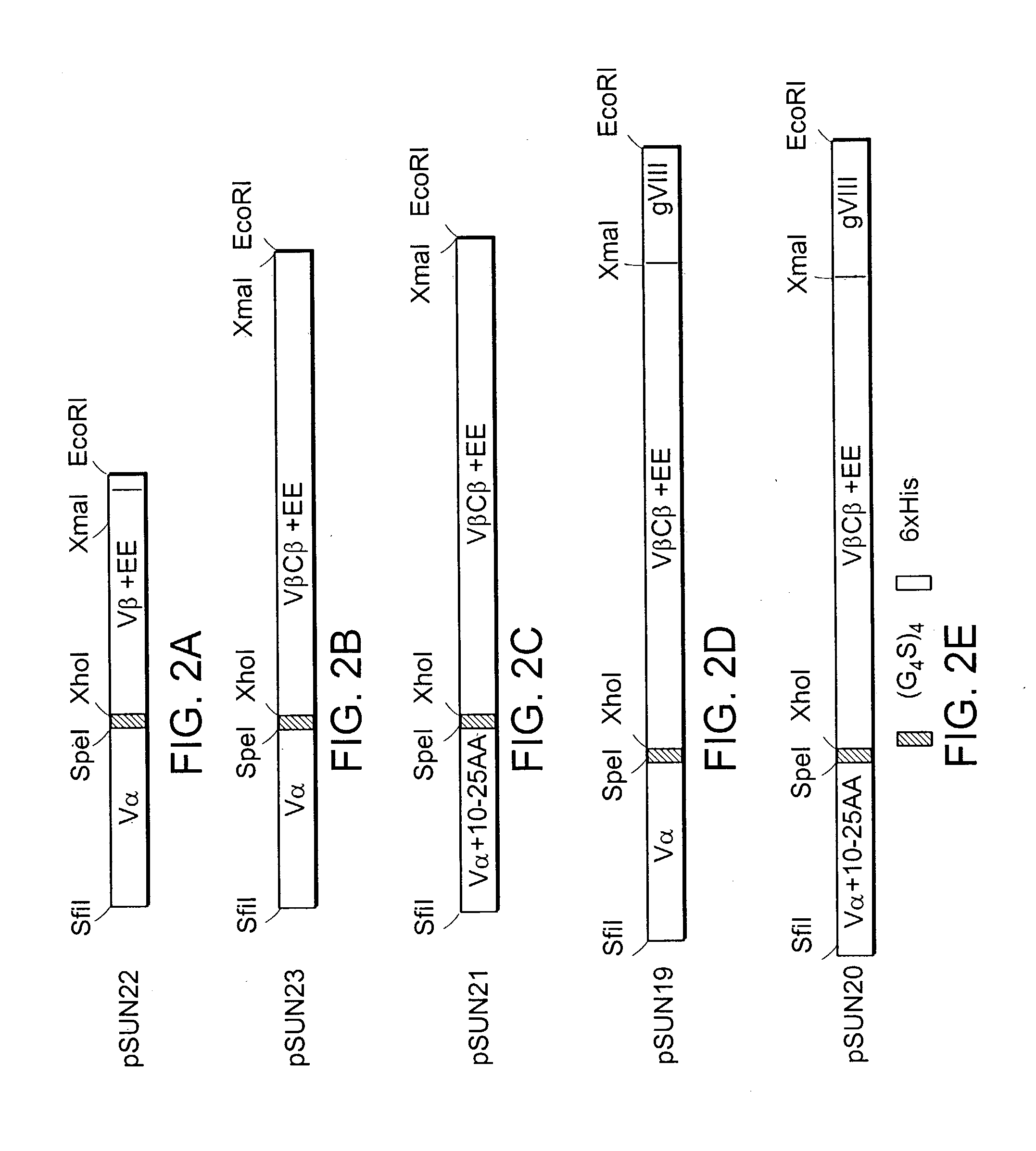
[0288] The DO11.10 TCR recognizes OVA peptide (323-339) in the context of the class II MHC IA.sup.d molecule. (See Haskins et al., J. Exp. Med., 1983.) The E. coli DNA construct pKC60 was reamplified by PCR with primers KC 169 and KC208 to generate a 5'AgeI-3'HpaI / BspEI / NruI / ClaI DNA fragment. The scTCR DNA fragment was cloned into the pGEM-T Easy Vector System for DNA sequence determination. The correct scTCR DNA was then restriction digested with AgeI and HpaI and cloned into the "shuttle vector", replacing the previous scTCR DNA fragment, to generate a new scTCR / scSc-Fv bispecific sc molecule. The DO11.10 bispecific sc molecule was then cloned into pSUN27 to create pBISP / DO11.10 (FIG. 6).
[0289] The pBISP / DO11.10 vector (PSUN 28) has been deposited pursuant to the Budapest treaty with the ATCC on Sep. 3, 1998 and was assigned Accession No. 203186.
[0290] 1. Expression of Variant scTCR Molecules in E. coli.
[0291] The ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| ionic strength | aaaaa | aaaaa |
| ionic strength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com