Methods for assaying gene imprinting and methylated cpg islands
a technology of methylation and cpg islands, which is applied in the direction of genetically modified cells, drug compositions, skeletal/connective tissue cells, etc., can solve the problems of hampered experimental studies of the timing and mechanism of genomic imprinting
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
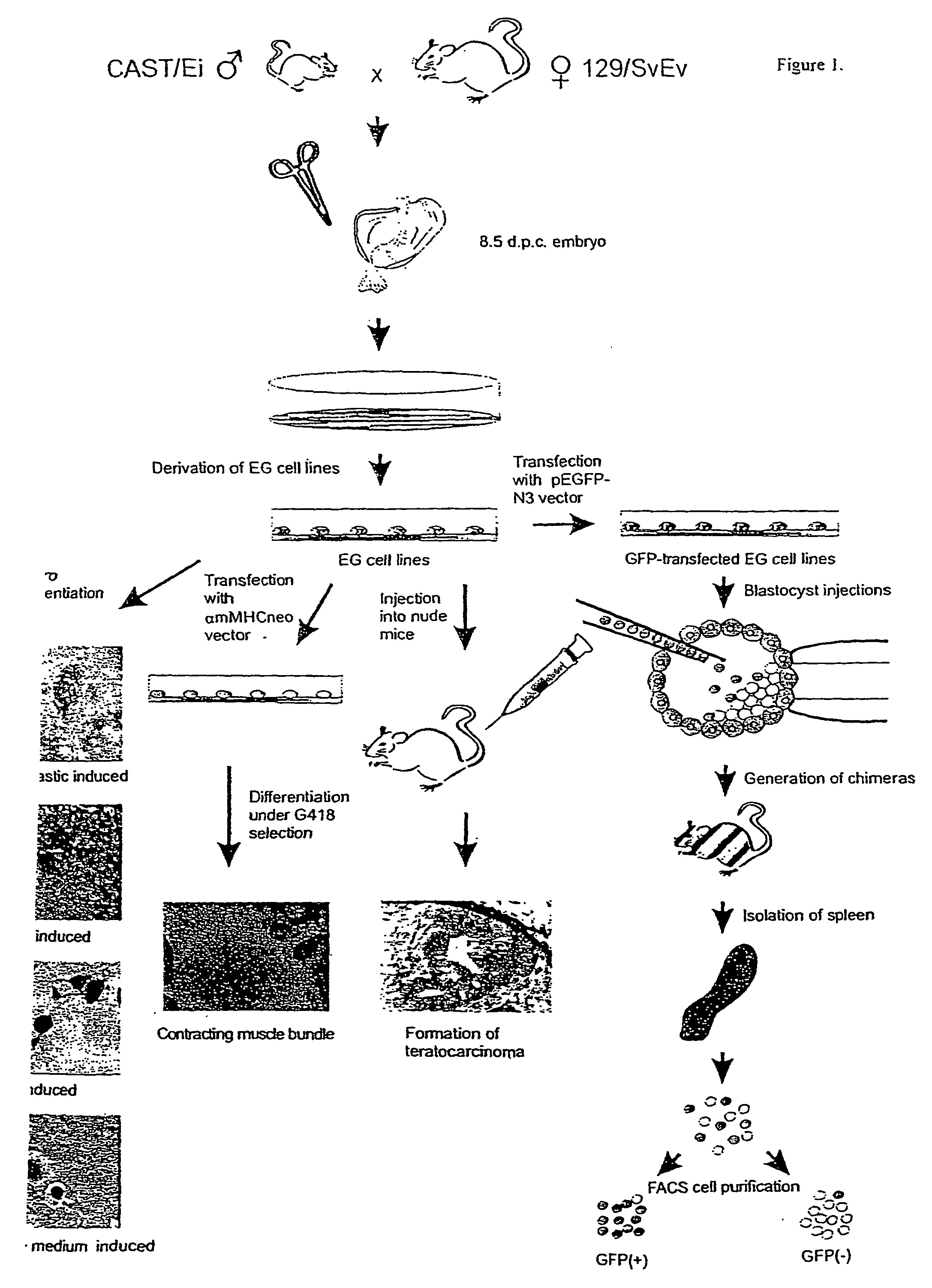
[0063] We used 129 / SvEv mice as the mothers in the cross We chose CAST / Ei (Mus musculus castaneus) mice, separated from 129 / SvEv by 5 million years in evolution, as the father in the cross, providing an average of one polymorphic marker per 400 bp of transcribed sequence. The experimental strategy is summarized in FIG. 1, and it allows differentiation in vitro by a variety of mechanisms, including targeted differentiation using a selectable construct, and differentiation in vivo using chimeric mice.
[0064] Forty EG cell lines were derived from primordial germ cells (PGCs) of 8.5 day embryos (4), as determined by colony morphology and positive alkaline phosphatase staining (FIG. 2A,B), and four of these lines were characterized in detail (termed SJEG-1, 2, 7, and 15). These EG cell lines formed embryoid bodies after in vitro differentiation (FIG. 2C,D), teratocarcinomas in nude mice (FIG. 2E, F), and generated chimeric mice when injected into the blastocyst of C57BL / 6 mice (5). One m...
example 2
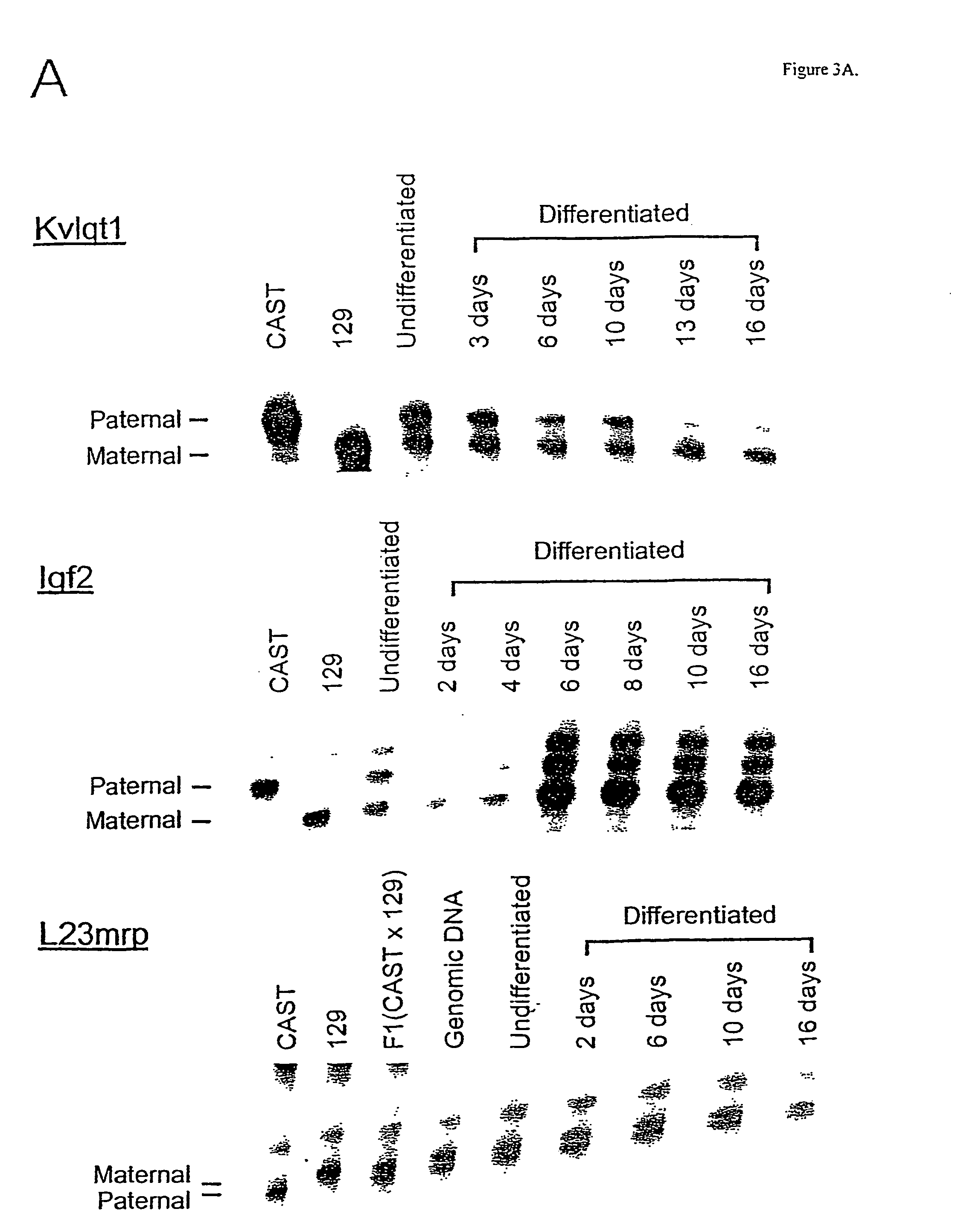
[0065] Partial establishment of imprinting in vitro. In order to distinguish the two alleles of imprinted genes in these EG cell lines, we identified transcribed polymorphisms distinguishing 129 / SvEv and CAST / Ei in 5 imprinted genes, Kvlqt1, Snrpn, Igf2, H19, and Igf2r, as well as the nonimprinted gene L23mrp as a negative control. For each gene, an assay for allele-specific expression was then developed, as described in Table 1.
TABLE 1Transcribed polymorphisms and assay methods forallele-specific gene expression of EG cellsderived from mouse interspecific cross.PolymorphismGeneCAST / Ei1129 / SvEvPosition2Assay MethodKvlqt1TCCCTGCTCCATGC1823SSCP3Igf2GCAATTCGCAGTTC777SSCP3H19CTTGGAGCTTTGAG1593QS4SnrpnCTATAATCTACAAT915SNuPE5Igf2rATCGATGATCAATG1549SNuPE5L23mrpACCCGAGACCTGAG407SSCP3
1Polymorphisms were identified by direct sequencing of CAST / Ei genomic DNA. 129 / SvEv sequence was identical to known Mus musculus musculus sequence in GenBank, except that Kvlqt1 sequence was unavailable and d...
example 3
[0069] Imprinting was independent of differentiation method. In order to determine whether allele-specific expression in EG cells was caused by differentiation in vitro, or by the specific treatment used to differentiate EG cells, we repeated these experiments by differentiating the cells in 3 other ways (4): differentiation in methylcellulose medium; treatment with retinoic acid; and treatment with dimethyl sulfoxide. In all cases, the results were identical to those seen on spontaneous differentiation on plastic in the absence of a feeder cell layer. For example, Snrpn showed equal biallelic expression of the two parental alleles prior to differentiation, and preferential expression of the paternal allele after differentiation in all cases, but with slight variation in the final ratio of parental alleles (FIG. 4A).
[0070] Embryoid bodies that result from in vitro differentiation of EG cells show considerable cellular heterogeneity, and not all of the cells are differentiated. In o...
PUM
| Property | Measurement | Unit |
|---|---|---|
| frequency | aaaaa | aaaaa |
| phase contrast microscopy | aaaaa | aaaaa |
| alkaline | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com