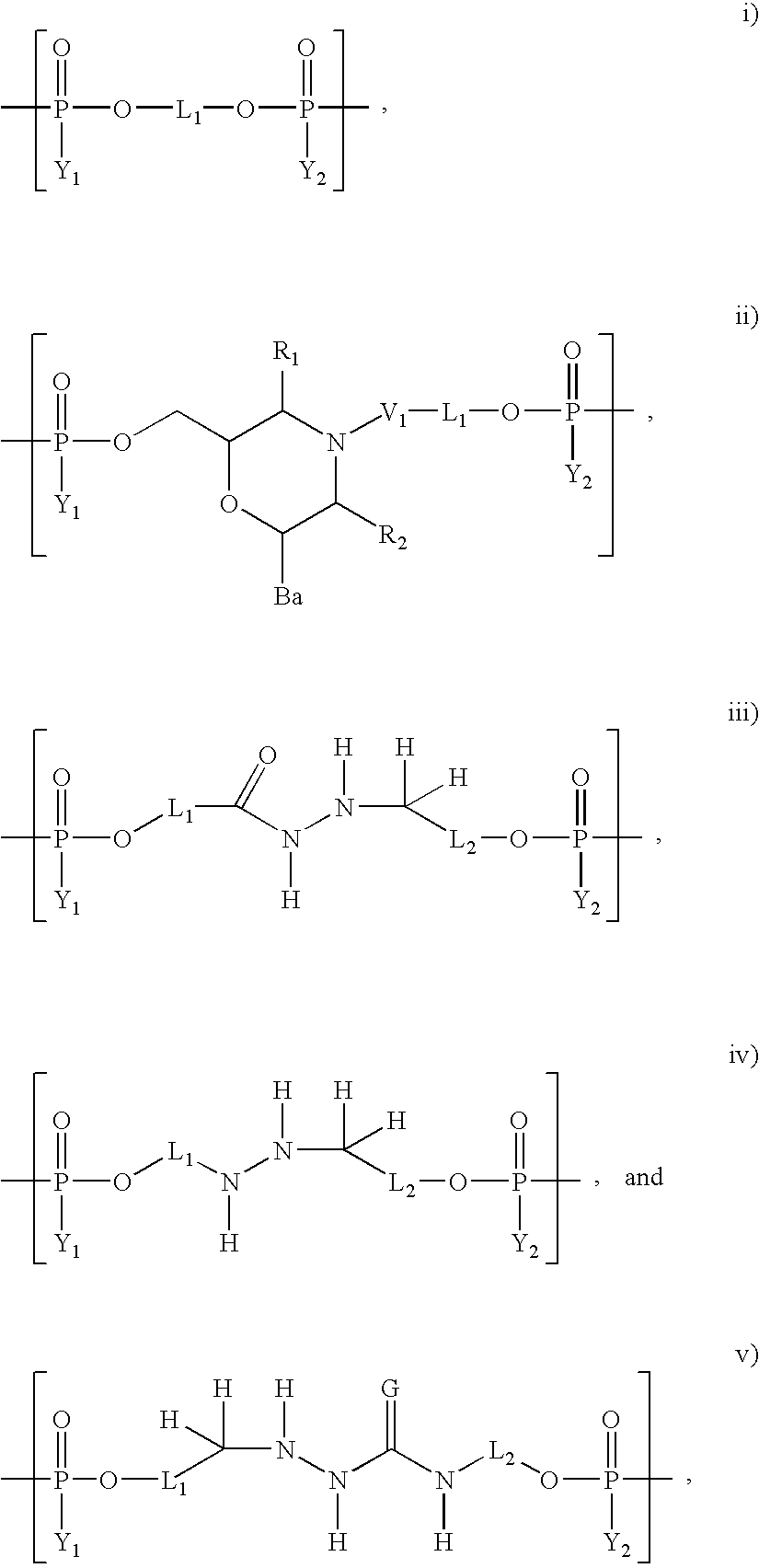
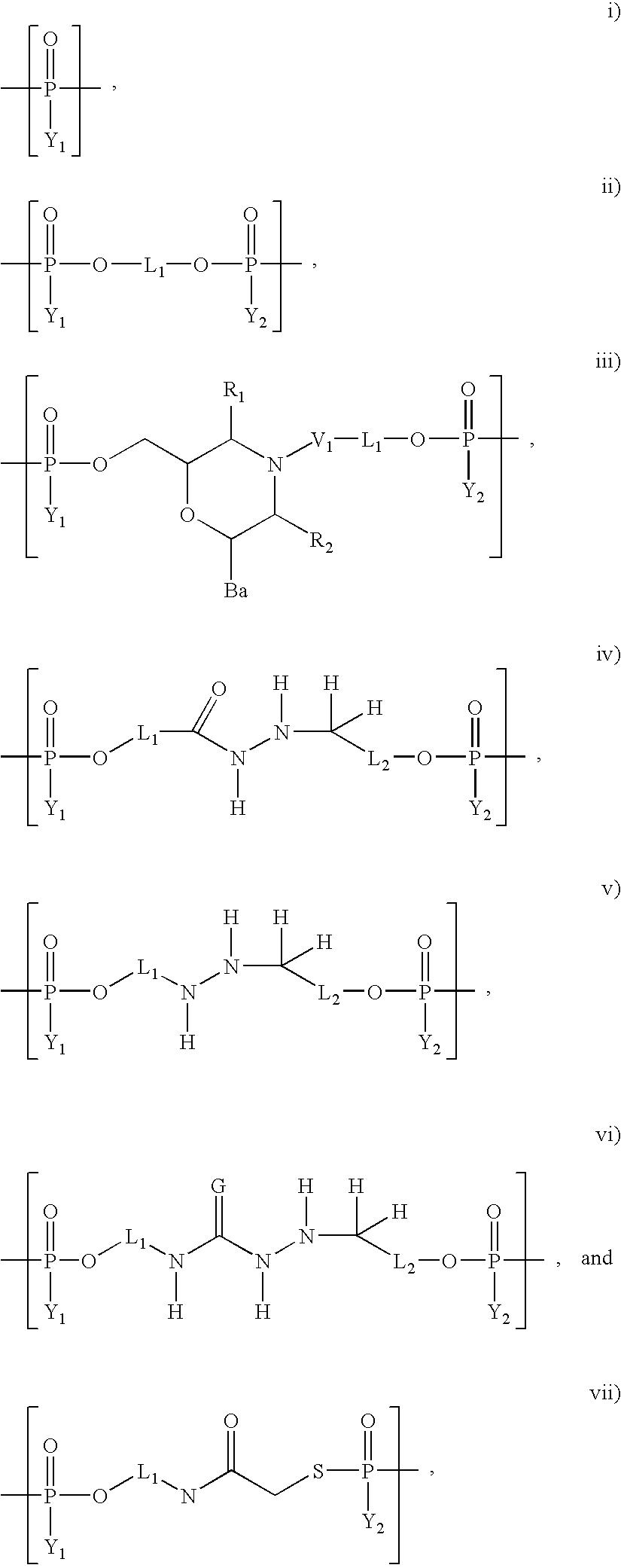
Sorting and immobilization system for nucleic acids using synthetic binding systems
a technology of synthetic binding and immobilization system, which is applied in the field of conjugating synthetic binding units and nucleic acids, can solve the problems of increasing the risk of unwanted or interfering interactions, increasing the complexity of samples, and potentially immobilizing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Synthesis of Conjugates of Nucleic Acids and Synthetic Binding Systems
[0230] Nucleic acids, oligonucleotides, synthetic binding systems and / or NA / SBU conjugates may be prepared according to the solid phase method on an automated synthesizer Expedite 8905 from Applied Biosystems. As utilized in the following examples, nucleic acids or oligonucleotides or the corresponding nucleic acid part of conjugates are synthesized using commercially available phosphoramidites including the reverse phosphoramidites for 5′-3′ synthesis (Cruachem). For solid synthesis of nucleic acids, the synthesis cycles and conditions suggested by the manufacturer of the equipment and / or amidite supplier were used. The same is true for the use of linking building blocks, linkers, modifiers or branching amidites (as available from Glenn Research or Chemgenes).
[0231] For pRNA synthetic binding systems, the conditions and monomers described in the appropriate references are used. Although the basic method for sol...
example 1.1
Conjugates of4′ pRNA2′-5′DNA3′ (FIG. 1C; FIG. 2C; FIG. 2F)
[0239] The synthesis was begun with a standard solid phase DNA synthesis in the 3′-5′ direction to produce the desired DNA sequence. If a linking group or branching group was desired, the appropriate synthesis building blocks were coupled to the 5′ end of the product under the conditions indicated by the manufacturer. If the DNA was directly linked to pRNA through a phosphate, (FIG. 2C), no linker phosphoramidite was used. Next, the monomer building blocks of pRNA were coupled under the special conditions described above so that a SBU binding address of the desired length and sequence was obtained. If a labeled conjugate was desired, marker phosphoramidites were coupled to the conjugate in an optional last step, or utilized as an initial coupled reagent in the synthesis. Here, the fluorescent dye phosphoramidites Cy3 and Cy5 (Amersham Pharmacia Biotech) were useful, but it is also possible to use any other suitable phosphora...
example 1.2
Conjugates of5′DNA3′-4′pRNA2′ (FIG. 1A: FIG. 2A: FIG. 2D)
[0241] The synthesis was begun with a pRNA synthesis as described above, producing the desired pRNA SBU on the synthesis support. If a linking group or branching group was desired, the appropriate synthesis building blocks were coupled to the product using conditions indicated by the manufacturer. As above, when pRNA was directly linked to DNA by a phosphate (FIG. 2A), no further linker was added. Standard DNA phosphoramidites and synthesis cycles were then used in order to synthesize the DNA sequence desired. If a labeled conjugate was desired, marker phosphoramidites were be coupled to the conjugate as described above. The conjugate was worked up, deprotected, isolated and purified as described above. Analogously, conjugates of 2′O-methyl RNA and pRNA were also obtained by using if 2′-O-methylphosphoramidites instead of DNA phosphoramidites.
[0242] Applying the general protocol led to the following conjugates (bold=pRNA; it...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com