Method for detecting acetyltransferase and deacetylase activities and method for screening inhibitors or enhancers of these enzymes
a technology which is applied in the field of acetyltransferase and deacetylase activities detection and screening methods for inhibitors or enhancers of these enzymes, can solve the problems of difficult assaying many samples under numerous conditions, cumbersome methods known for measuring acetyltransferase and deacetylase activities, etc., and achieves convenient screening and convenient detection of a
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
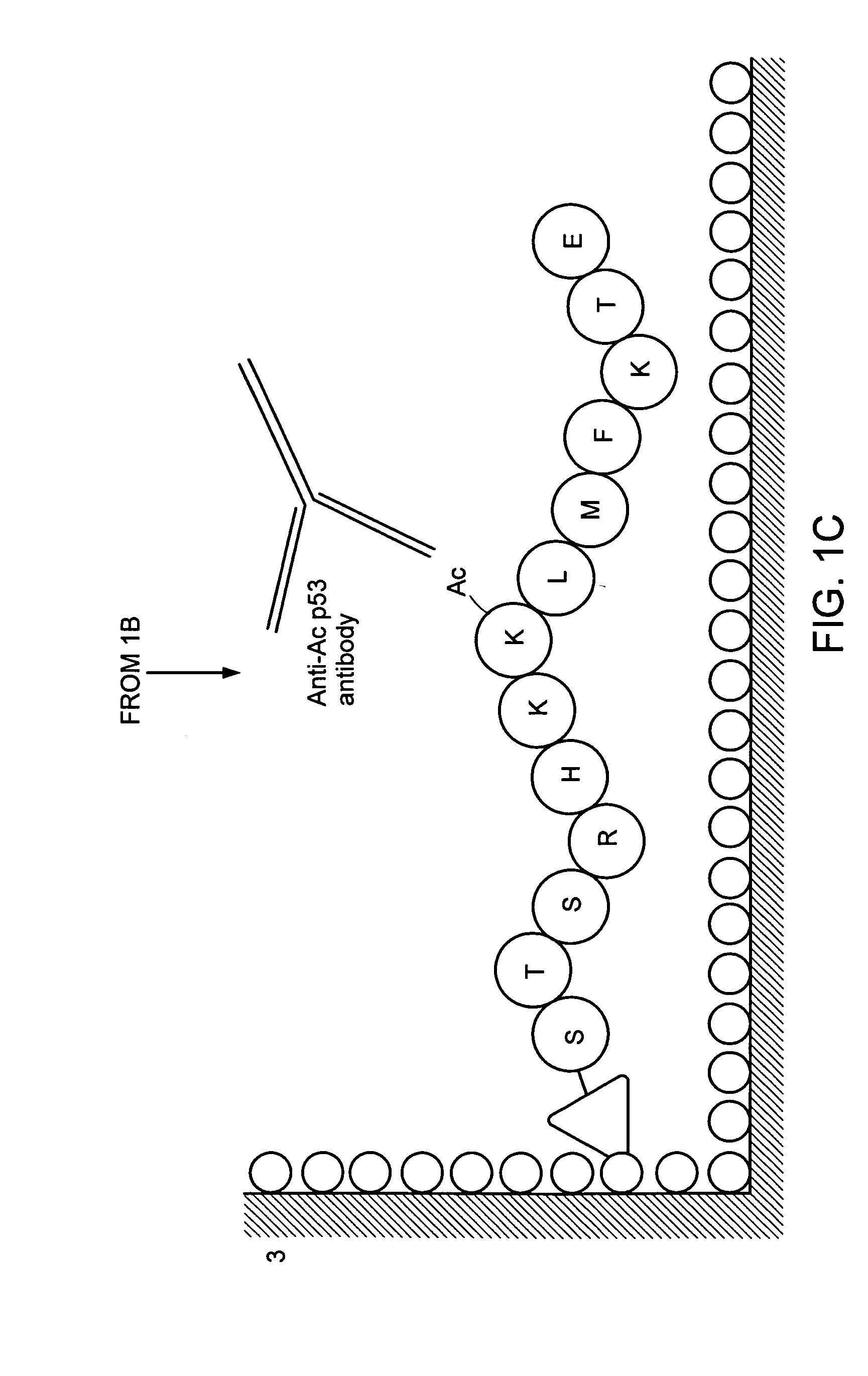
Preparation of an Anti-Acetylated Peptide Antibody
1. Preparation of an Immunogen
(1) Preparation of a Peptide
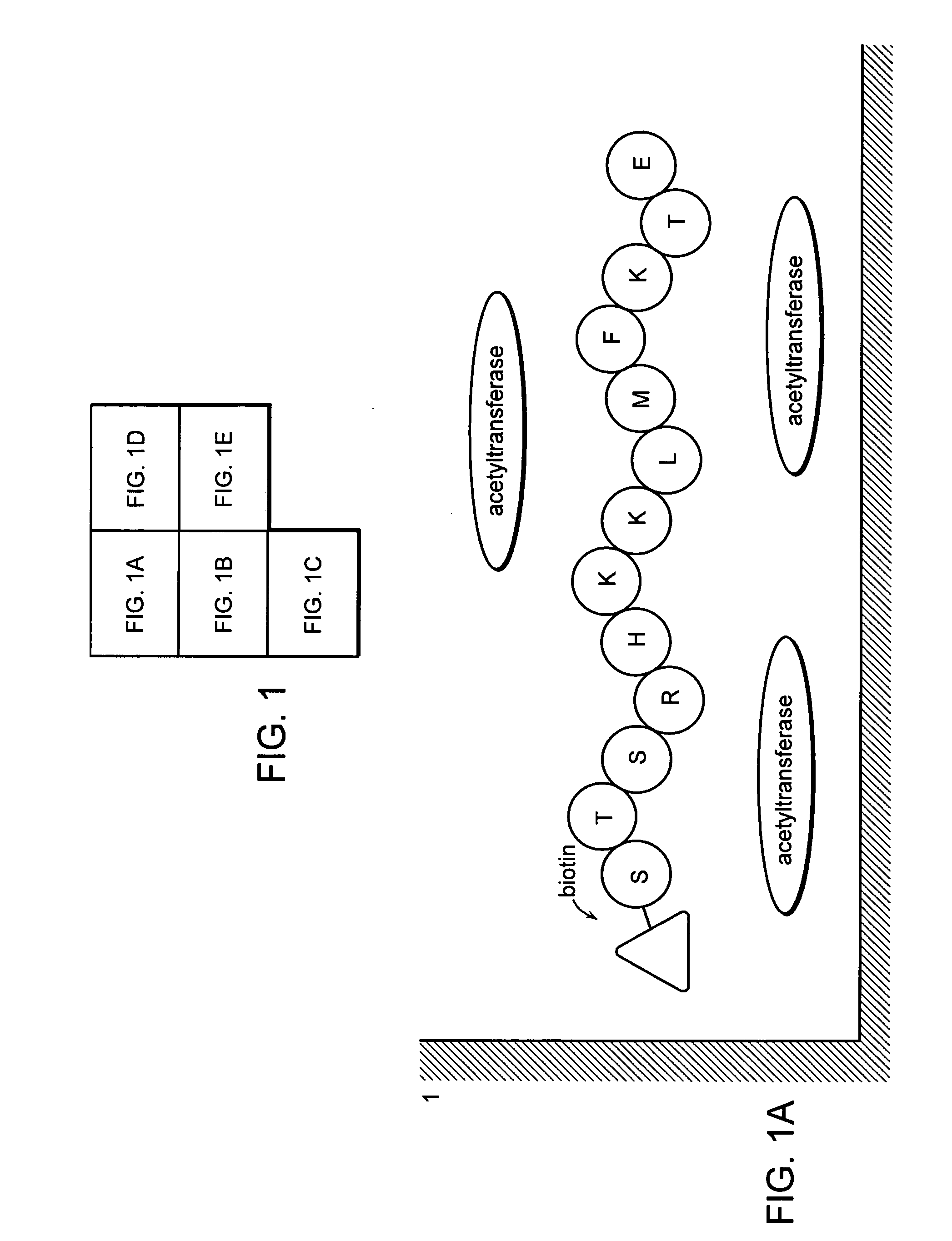
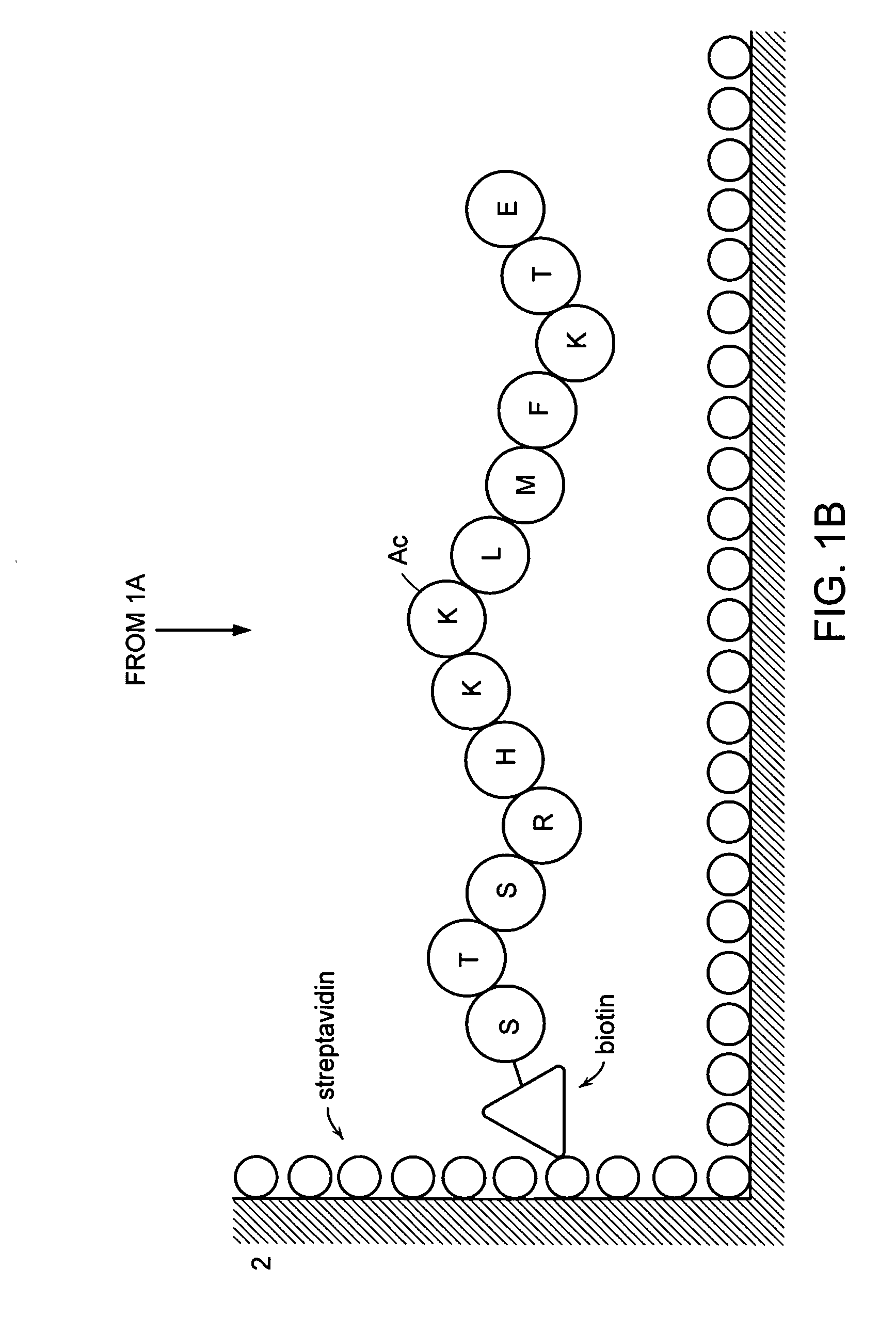
[0103] Four peptides comprising the 373rd and 382nd lysine residues of human p53, reported as acetylation sites were prepared by a peptide synthesizer. These peptides are “Ac p53-1” (SEQ ID NO: 1 / STSRHKK (Ac) LMFKTEC), “p53-1” (SEQ ID NO: 2 / STSRHKKLMFKTEC), “Ac p53-2” (SEQ ID NO: 3 / SHLKSKK (Ac) GQSTSRC) and “p53-2” (SEQ ID NO: 4 / SHLKSKKGQSTSRC). Amino acids are given in one-letter codes, and K(Ac) indicates an acetylated lysine residue. HPLC confirmed that the purity of synthesized peptides was 90% or more. “Ac p53-1” and “p53-1” are composed of amino acids residues 367 to 388 of human p53, “Ac p53-2” and “p53-2” are composed of amino acid residues 367 to 379 of human p53. The cysteine residue at the carboxyl terminus in all peptides was inserted for binding a peptide into a carrier protein.
(2) Binding a Peptide into a Carrier Protein
[0104] Each acetylated peptide (“Ac...
example 2
Preparation of Acetyltransferase and Deacetylase by Genetic Engineering
1. Isolation of Genes by PCR
[0112] Up to now, as acetyltransferases of p53 and mammalian histone, five genes: P300 / CBP, Gcn5, TAFII250, P / CAF, and Tip 60 have been reported. Among them, P300 and CBP are different genes, however, their nucleotide and amino acid sequences are highly homologous. Among these acetyltransferases, P300 / CBP and Gcn5 were amplified and isolated by PCR method. The following PCR primers were prepared. For amplifying P300 / CBP, a forward primer “CBPF” [SEQ ID NO: 5 / 5′-GCGGGATCCCAGAATAGGTATCATTTCTGTGAG-3′ [the three nucleotides (GCG) at the 5′ end were added for enhancing the treatment with restriction enzymes, and forth to ninth nucleotides (GGATCC) from the 5′ end is the restriction enzyme BamHI site] and a reverse primer “CBPR” [SEQ ID NO: 6 / 5′-AGACTCGAGCTTGCACTCGTTGCAGGTGTAGAC-3′ (the three nucleotides (AGA) at the 5′ end were added for enhancing the treatment wit...
example 3
System for Measuring the Acetyltransferase Activity
1. Construction of an ELISA System for Measuring the Acetyltransferase Activity
(1) Construction of a Peptide Substrate and Recombinant p53-C ter Protein
[0122] Two peptide substrates “Sub p53-1” (SEQ ID NO: 13 / Bio-STSRHKKLMFKTE) and “Sub p53-2” (SEQ ID NO: 14 / Bio-SHLKSKKGQSTSR) comprising the 373rd and 382nd lysine residues of the amino acid sequence of human p53, respectively, reported as an acetylation sites of acetyltransferase, were prepared by a peptide synthesizer. Amino acids in the peptide are shown in one-letter codes, and “Bio” at the amino ends means biotin. Ninety percent or higher purity was confirmed by HPLC. “Sub p53-1” and “Sub-p53-2” were composed of amino acids 376 to 388 and amino acids 367 to 379, respectively. The DNA comprising the genetic information of 110 amino acids (residues 284 to 393) at the carboxyl end in p53 was amplified using the primer set “p53cF” [SEQ ID NO: 15 / 5′-TATGGATCCACAGAGGAAGAGAATCTCCG...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com