Prevention of disulfide bond reduction during recombinant production of polypeptides
a technology of disulfide bond and recombinant protein, which is applied in the direction of peptides, drug compositions, immunological disorders, etc., can solve the problems that the production of recombinant proteins is still not without difficulties, and achieve the effect of preventing the reduction of a disulfide bond
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Description of Materials and Methods
[0281]The following materials and methods were used in Examples 2-8 below.
[0282]Materials
[0283]Materials and devices used in the experiments described in the experimental examples include: stainless steel vials (mini-tanks, Flow Components, Dublin, Calif.; short (50 cc) and tall (55 cc)); dialysis tubing (Spectra / Por, 6-8000 MWCO, cat. #132645), 0.22 μm filter (Millipore Millipak Gamma Gold cat. #MPGL04 GH2); phosphate buffered saline (PBS, EMD, cat. #6506); ethylenediaminetetraacetic acid (EDTA, Sigma, cat. #E4884); α-nicotinamide adenine dinucleotide phosphate (NADPH, Calbiochem, cat. #481973); dehydroepiandrosterone (DHEA, TCI, cat. #D0044); cupric sulfate (Sigma, cat. #C8027), glucose-6-phosphate (G6P, Calbiochem, cat. #346764); aurothioglucose (ATG, USP, cat. #1045508); aurothiomalate (ATM, Alfa Aesar, cat. #39740); reduced glutathione (GSH, J. T. Baker, cat. #M770-01); monobromobimane (mBB, Fluka, cat. #69898); histidine (J. T. Baker, cat. #...
example 2
Dialysis Experiment
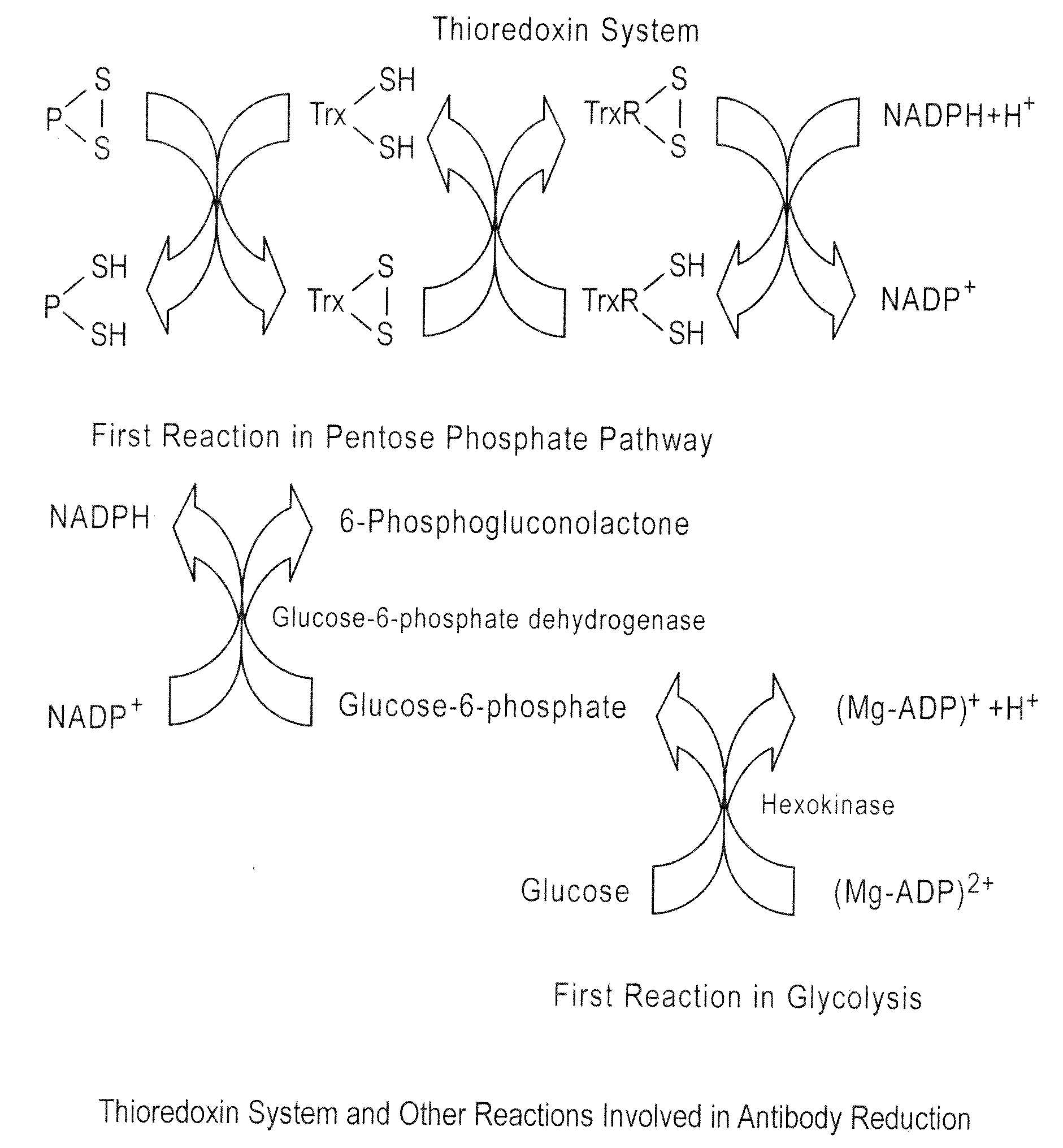
[0317]A dialysis experiment was designed and carried out to determine if the reduction of ocrelizumab was caused by small reducing molecules or macromolecules (e.g., enzymes). In this dialysis experiment, purified intact ocrelizumab was placed in a dialysis bag with a molecular weight cut off (MWCO) of 7000 and incubated the dialysis bag in HCCF containing ocrelizumab in a stainless steel mini-tank. As shown in FIGS. 1 and 2, the ocrelizumab inside the bag was not reduced after the incubation period (FIG. 1), whereas the ocrelizumab outside the bag in the HCCF was significantly reduced soon after the incubation started. This was evidenced by the loss of intact ocrelizumab (˜150 kDa) and the formation of ocrelizumab fragments (various combinations of heavy and light chains) (FIG. 2). The mass spectrometry analysis of the ocrelizumab in the protein A elution pools from the reduced manufacturing runs indicated that those observed fragments were formed by reduction of...
example 3
Reduction of Ocrelizumab (rhuMAb 2H7, Variant A) by Trx / TrxR In Vitro
[0319]The Trx system was tested for its ability to reduce ocrelizumab in vitro by incubating intact ocrelizumab with Trx, TrxR, and NADPH. The Bioanalyzer results indicate that ocrelizumab was reduced in vitro by the Trx system (FIG. 5). The rate of reduction in this in vitro system appears to be slower than that in the HCCF (for example when compared to the reduction shown in FIG. 2). This is likely due to lower concentrations of the enzymes (Trx and Trx-R) and / or the buffer system used in the in vitro reaction because reaction rate of Trx system is dependent on both the enzyme concentrations and buffer systems.
PUM
| Property | Measurement | Unit |
|---|---|---|
| hold time | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com