Systems and Methods to Quantify and Amplify Both Signaling and Probes for CDNA Chips and Gene Expression Microarrays
a technology of cdna chips and probes, applied in the field of detecting genes and gene expression from biological and medical samples, can solve the problems of ineffectiveness, inability to detect genes, and high cost of approach, and achieve the effect of improving diagnostic value and increasing signaling
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Sample Molecular Compositions of the Present Invention
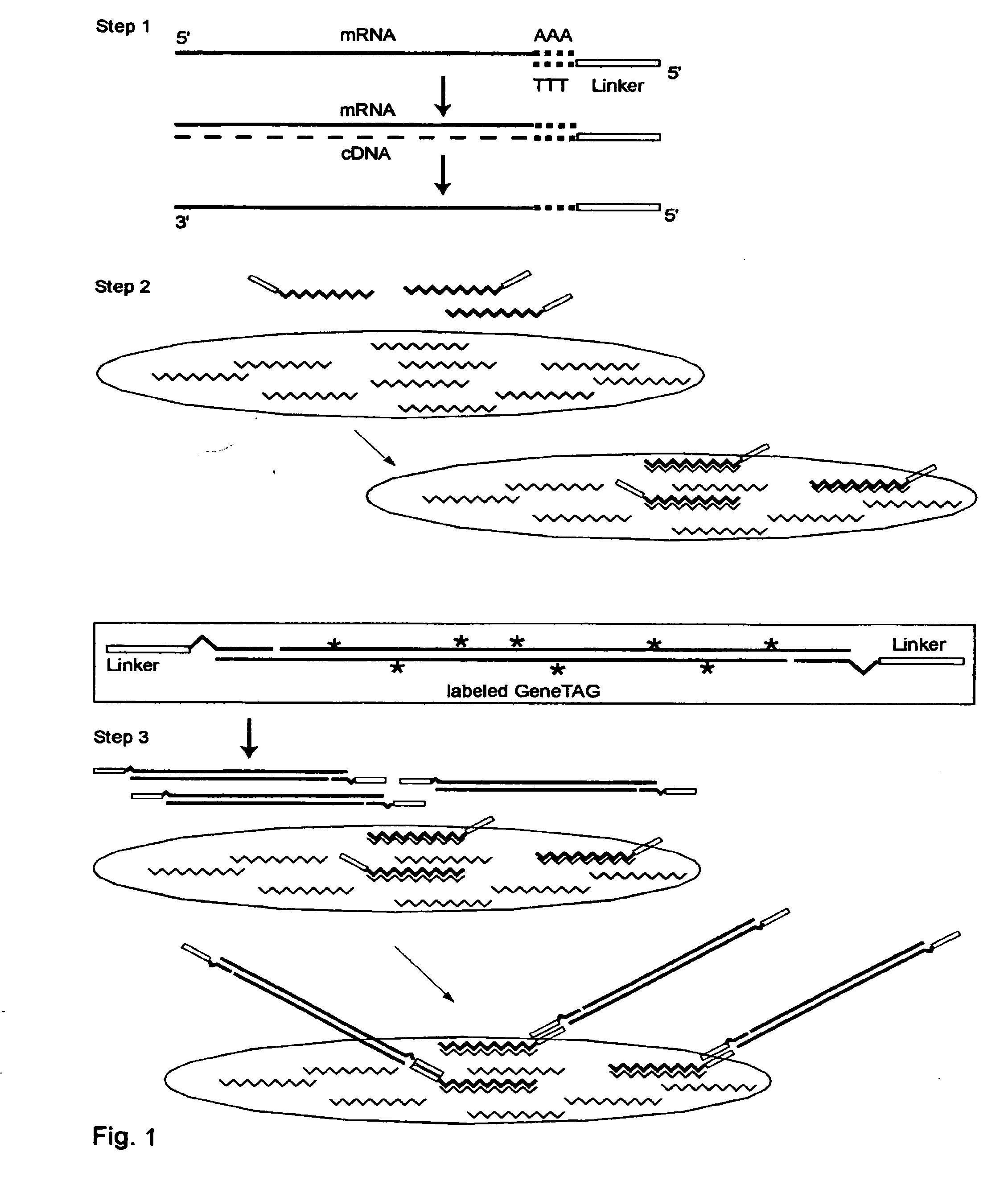
[0219]Universal GeneTAG Linkers:
[0220]1. Red 5′CTACGATACGATAGCGCCTAAGAGTAG (Seq. ID. No. 1) and its complement.
[0221]2. Green 5′CCTAGACCTACGACATAGGTACCCTAC (Seq. ID. No. 2) and its complement.
[0222]3. Blue 5′CGTAGAACTAGCACGCTACGTACTAGG (Seq. ID. No. 3) and its complement.
[0223]4. Orange 5′GGCTATCGCTACGTAGACTAGACCTAC (Seq. ID. No. 4) and its complement.
[0224]Modified Poly-T Primer with red or green universal linker and anchor end:
(Seq. ID. No. 1, 5)1. 5′CTACGATACGATAGCGCCTAAGAGTAG-TTTTTTTTTTTTTTTVN(Seq. ID. No. 2, 5)2. 5′CCTAGACCTACGACATAGGTACCCTAC-TTTTTTTTTTTTTTTVN
[0225]Double-Linker WRAP-Probe Set 1: Showing one probe strand with Red and Blue Universal Linkers, and with the variable target sequence indicated by S1 . . . Sn (Seq. ID. No. 1, 6)
[0226]5′CTACGATACGATAGCGCCTAAGAGTAG-S1 . . . Sn-CCTAGTACGTAGCGTGCTAGTTCTACG
[0227]Double-Linker WRAP-Probe Set 2: Showing one probe strand with Green and Orange Universal Linkers, and with th...
example 2
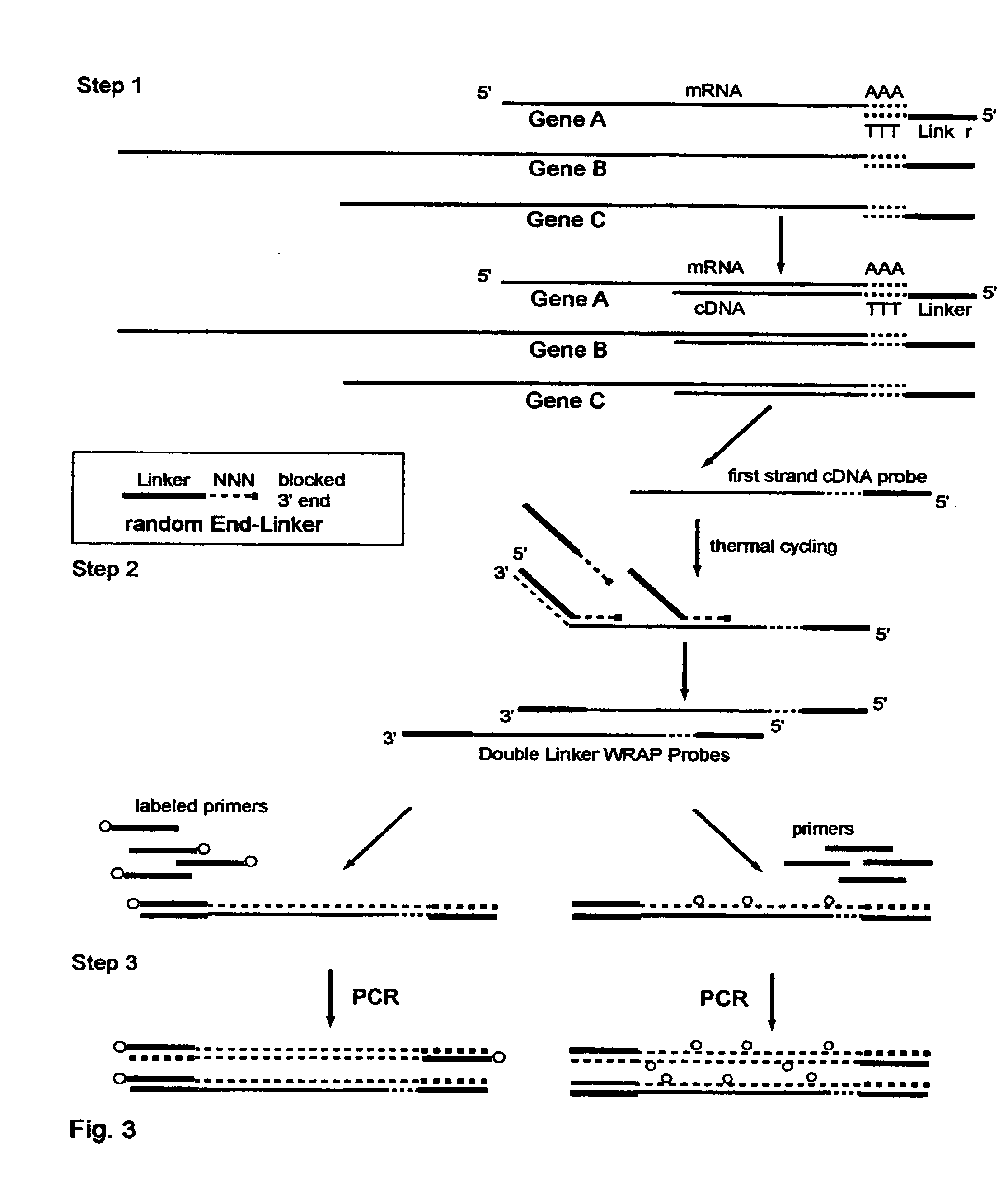
One-Linker WRAP-Probe Method
[0247]Total RNA is extracted from A549 lung cancer cells by standard methods. Reverse transcriptase (RT) is then employed to copy the mRNA transcripts to cDNA using a Modified poly-T Primer known as R-GT-RTP (Seq. ID. No. 8) having a 3′ end of 15 poly-T's and a 5′ end with a universal linker sequence that is similar to but differing in part from the Red Universal Linker of Example 1. For a comparative sample an alternative Modified poly-T Primer, known as G-GT-RTP (Seq. ID. No. 9) is used for the RT reaction to provide a second universal linker sequence wherein those sequences are also similar to but differing in part from the Green universal linker of Example 1.
[0248]The Examples 2 through 7 use these earlier versions of the red and green universal linkers in all their products, and thus the text of those Examples identifies them differently with the terms First-RED and First-GREEN in the descriptions to identify these sequence differences.
[0249]For a 25...
example 3
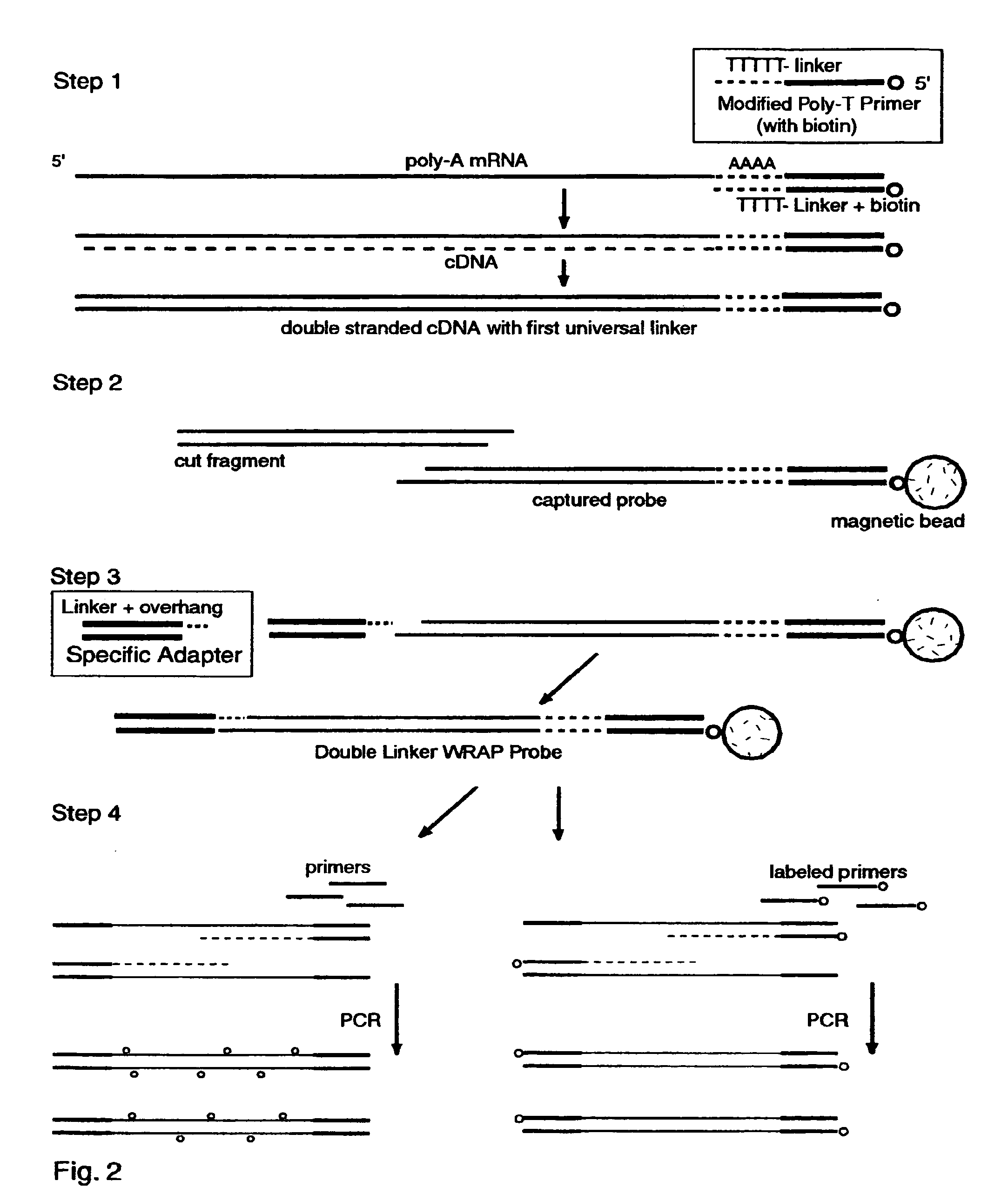
Double-linker WRAP-Probe Method with Restriction Cutting and Adapter Ligation
[0253]Total RNA was extracted from A549 lung cancer cells by standard methods. A 40 ug sample was treated with reverse transcriptase (RT) and a Modified Poly-T Primer to make ds cDNA copies of the mRNAs with a first linker, to cut and capture the end fragments, and to add a second linker by ligating an adapter. The following steps were employed to make the probes and perform a chip analysis:
[0254]1. Full length first strand cDNAs were made with one hour exposure to MuVL RT (Gibco) at 37 degrees C. using a Modified Poly-T Primer (Seq. ID. No. 16) having a 5′ biotin capture moiety, an overlap linker sequence and a poly-T segment. Nucleotides including Cy3-dCTP (AP Biotech) were incorporated as described above to provide “green” labeling.
[0255]2. Double stranded cDNA was made with E. coli DNA polymerase I, Rnase H and DNA ligase (Gibco kit) with a two hour exposure at 16 degrees C.
[0256]3. The ds cDNAs were tr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com