Recombinational cloning using nucleic acids having recombination sites
a nucleic acid and recombination technology, applied in the field of recombinant dna technology, can solve the problems of high background, toxic genes, long fragments, etc., and achieve the effects of less labor, high specificity, speed and yield, and improved specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Recombinational Cloning Using Cre and Cre & Int
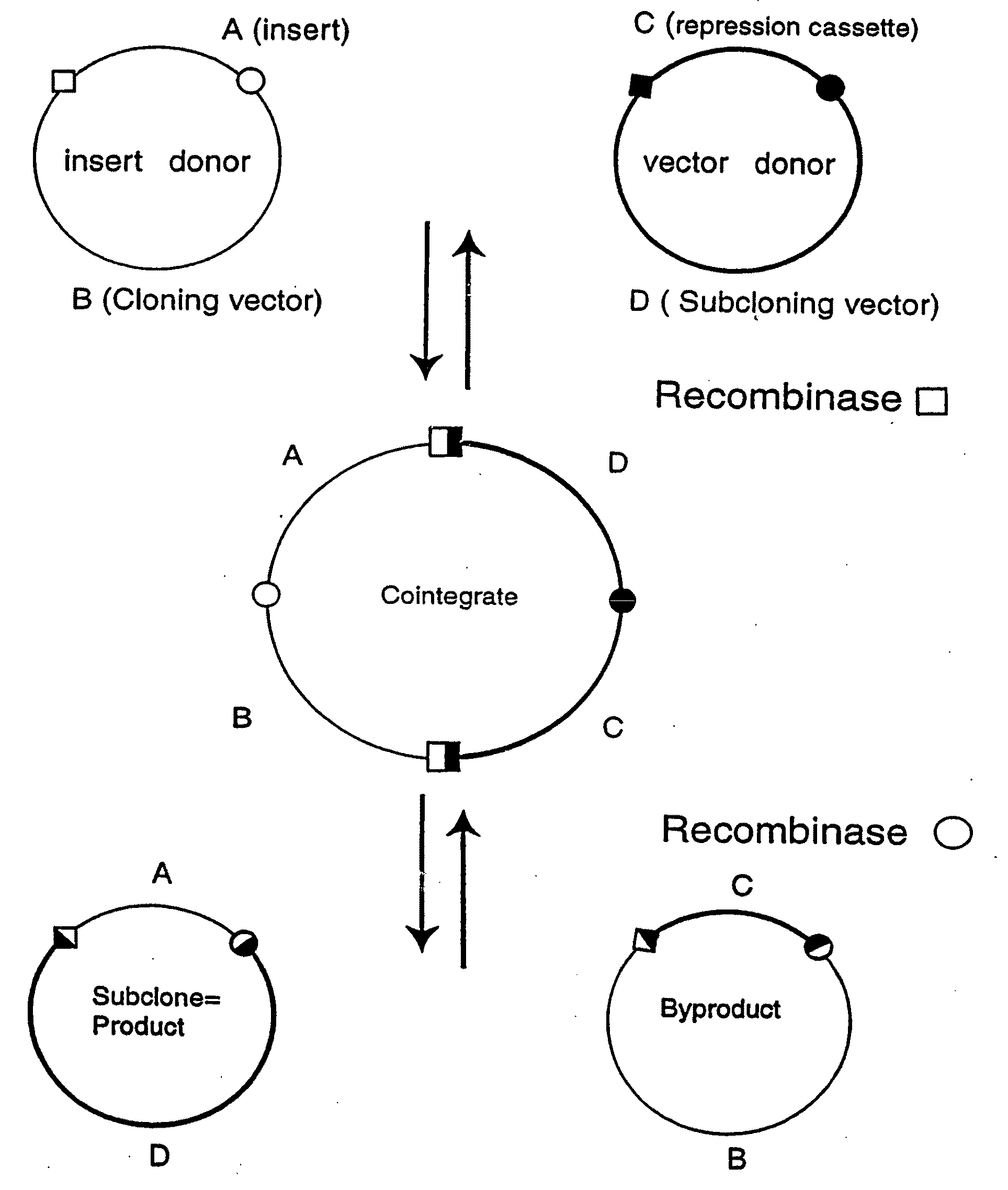
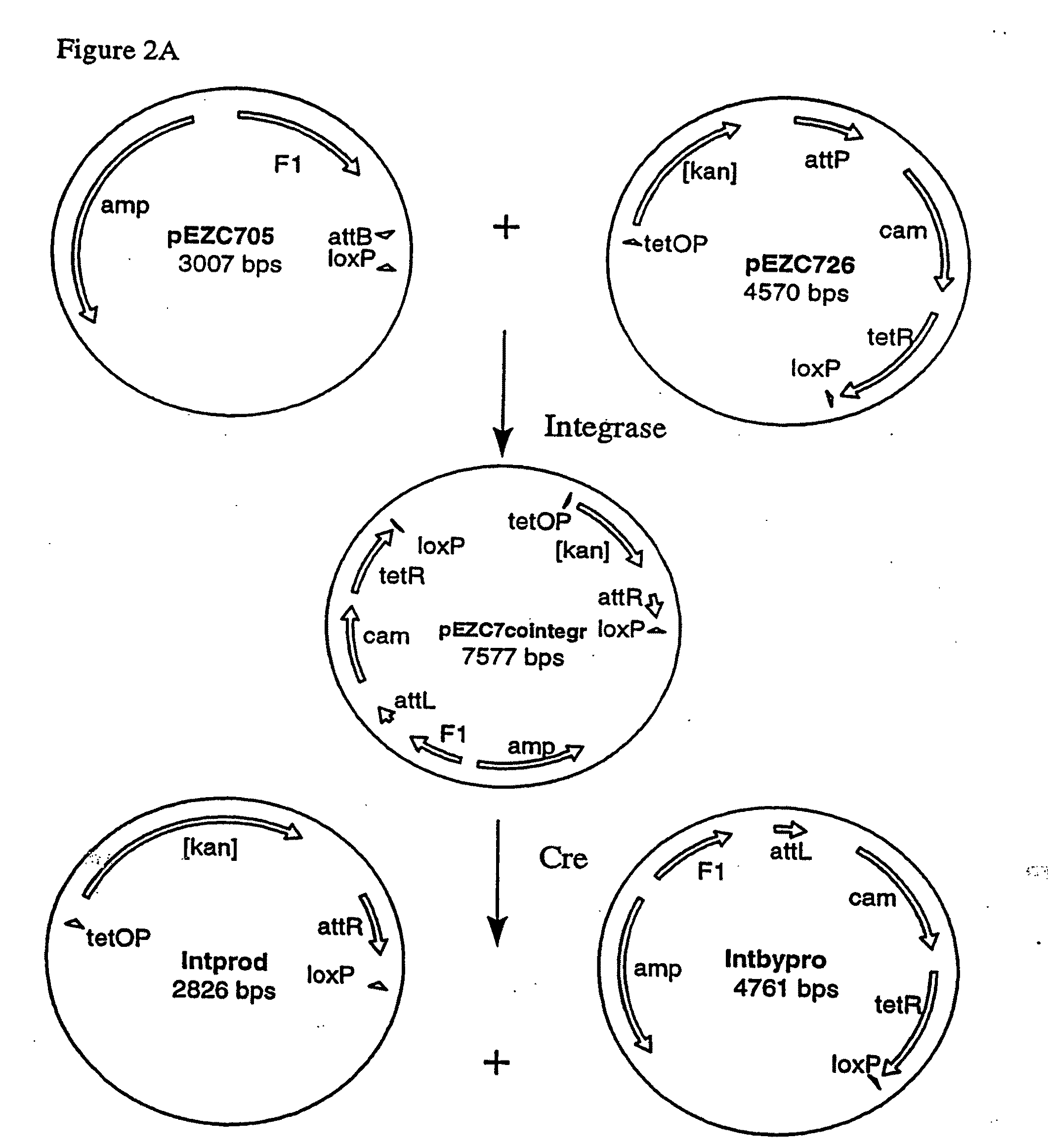
[0252]Two pairs of plasmids were constructed to do the in vitro recombinational cloning method in two different ways. One pair, pEZC705 and pEZC726 (FIG. 2A), was constructed with loxP and att sites, to be used with Cre and λ integrase. The other pair, pEZC602 and pEZC629 (FIG. 3A), contained the loxP (wild type) site for Cre, and a second mutant lox site, loxP 511, which differs from loxP in one base (out of 34 total). The minimum requirement for recombinational cloning of the present invention is two recombination sites in each plasmid, in general X and Y, and X′ and Y′. Recombinational cloning takes place if either or both types of site can recombine to form a Cointegrate (e.g. X and X′), and if either or both can recombine to excise the Product and Byproduct plasmids from the Cointegrate (e.g. Y and Y′). It is important that the recombination sites on the same plasmid do not recombine. It was found that the present recombinational c...
example 2
Using In Vitro Recombinational Cloning to Subclone the Chloramphenicol Acetyl Transferase Gene into a Vector for Expression in Eukaryotic Cells (FIG. 4A)
[0263]An Insert Donor plasmid, pEZC843, was constructed, comprising the chloramphenicol acetyl transferase gene of E. coli, cloned between loxP and attB sites such that the loxP site was positioned at the 5′-end of the gene (FIG. 4B). A Vector Donor plasmid, pEZC1003, was constructed, which contained the cytomegalovirus eukaryotic promoter apposed to a loxP site (FIG. 4C). One microliter aliquots of each supercoiled plasmid (about 50 ng crude miniprep DNA) were combined in a ten microliter reaction containing equal parts of lambda integrase buffer (50 mM Tris-HCl, pH 7.8, 70 mM KCl, 5 mM spermidine, 0.5 mM EDTA, 0.25 mg / ml bovine serum albumin) and Cre recombinase buffer (50 mM Tris-HCl, pH 7.5, 33 mM NaCl, 5 mM spermidine, 0.5 mg / ml bovine serum albumin), two units of Cre recombinase, 16 ng integration host factor, and 32 ng lambda...
example 3
Subcloned DNA Segments Flanked by AttB Sites without Stop Codons
Part I: Background
[0264]The above examples are suitable for transcriptional fusions, in which transcription crosses recombination sites. However, both attR and loxP sites contain multiple stop codons on both strands, so translational fusions can be difficult, where the coding sequence must cross the recombination sites, (only one reading frame is available on each strand of loxP sites) or impossible (in attR or attL).
[0265]A principal reason for subcloning is to fuse protein domains. For example, fusion of the glutathione S-transferase (GST) domain to a protein of interest allows the fusion protein to be purified by affinity chromatography on glutathione agarose (Pharmacia, Inc., 1995 catalog). If the protein of interest is fused to runs of consecutive histidines (for example His6), the fusion protein can be purified by affinity chromatography on chelating resins containing metal ions (Qiagen, Inc.). It is often desirab...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| total volume | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com