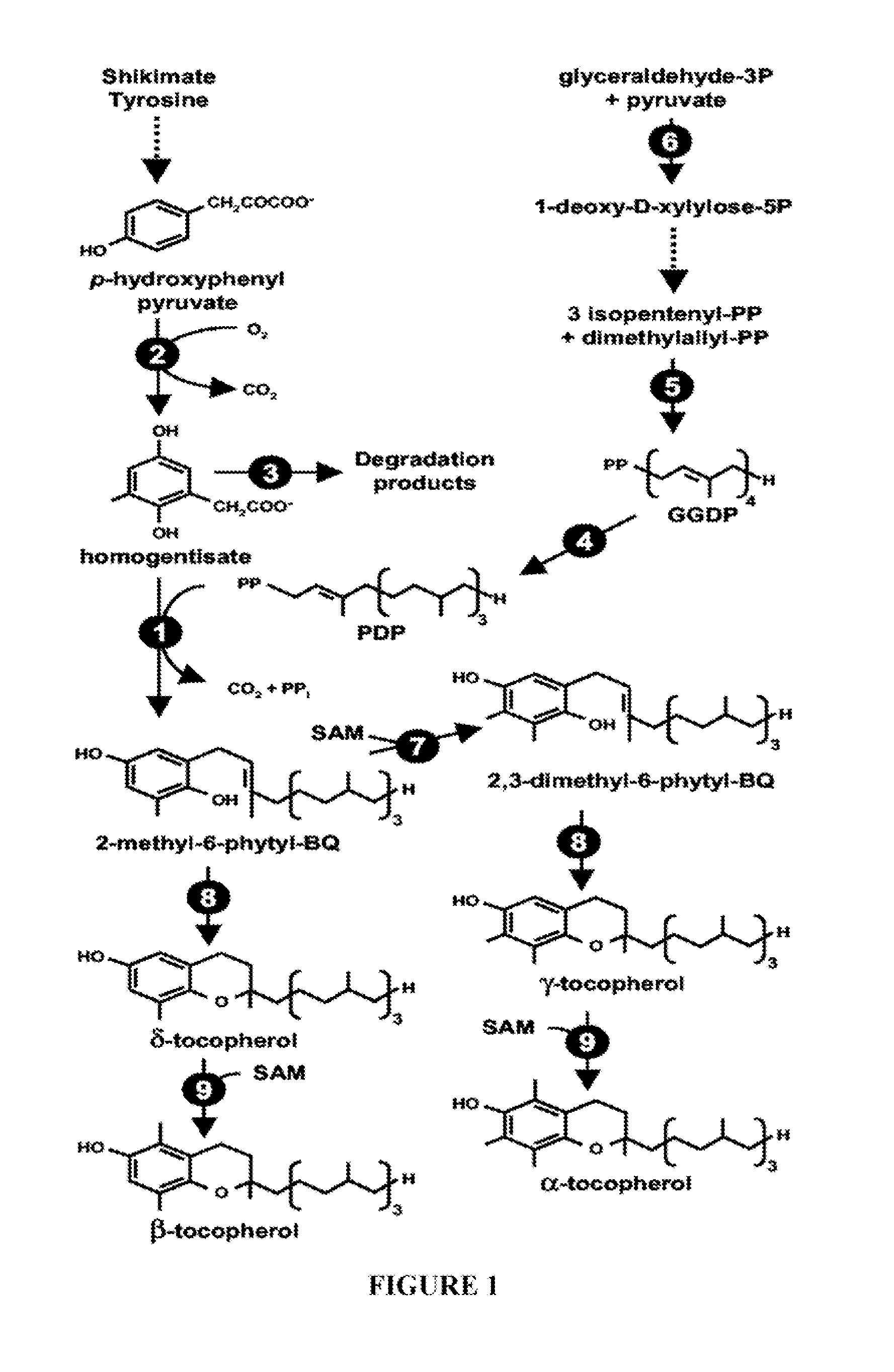
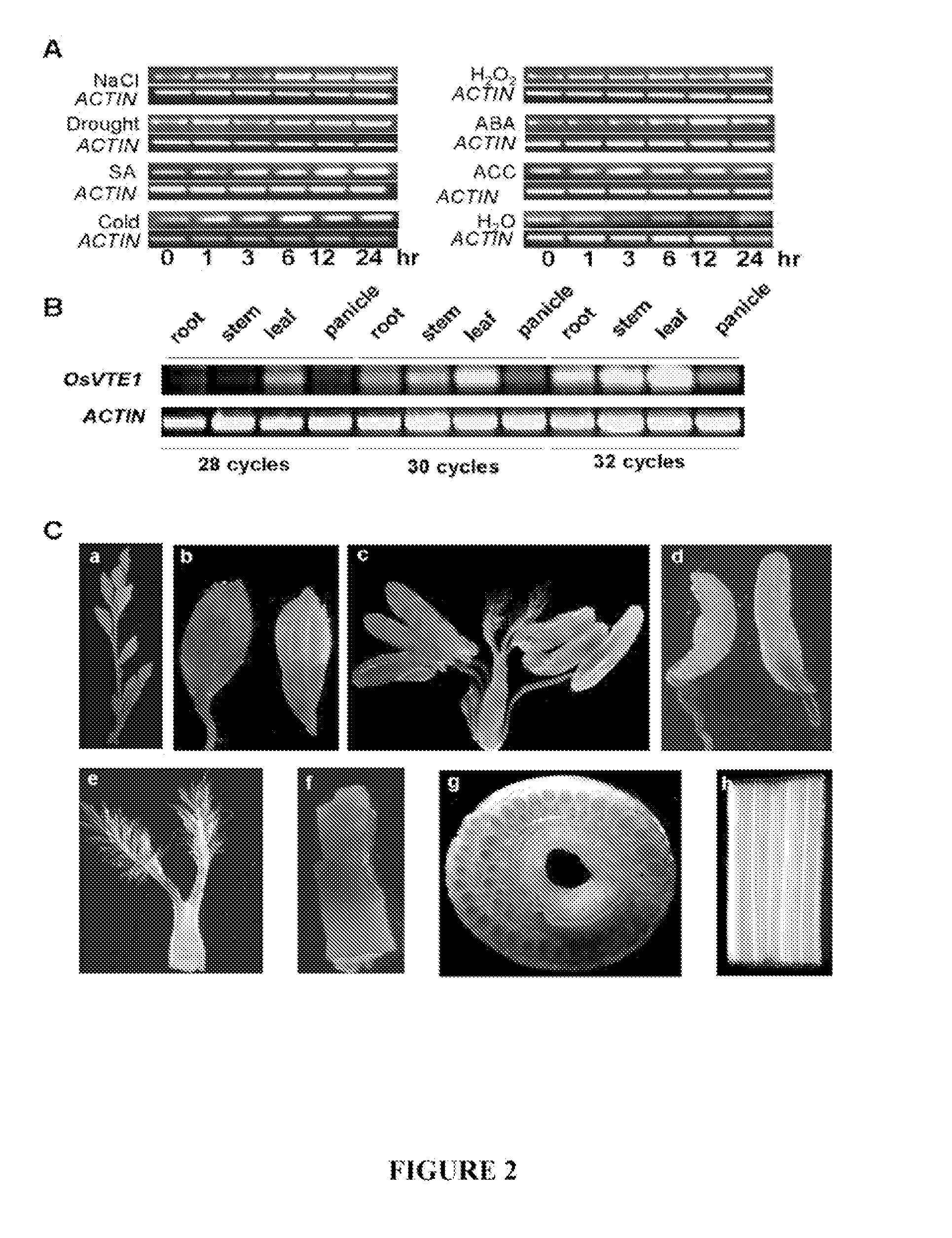
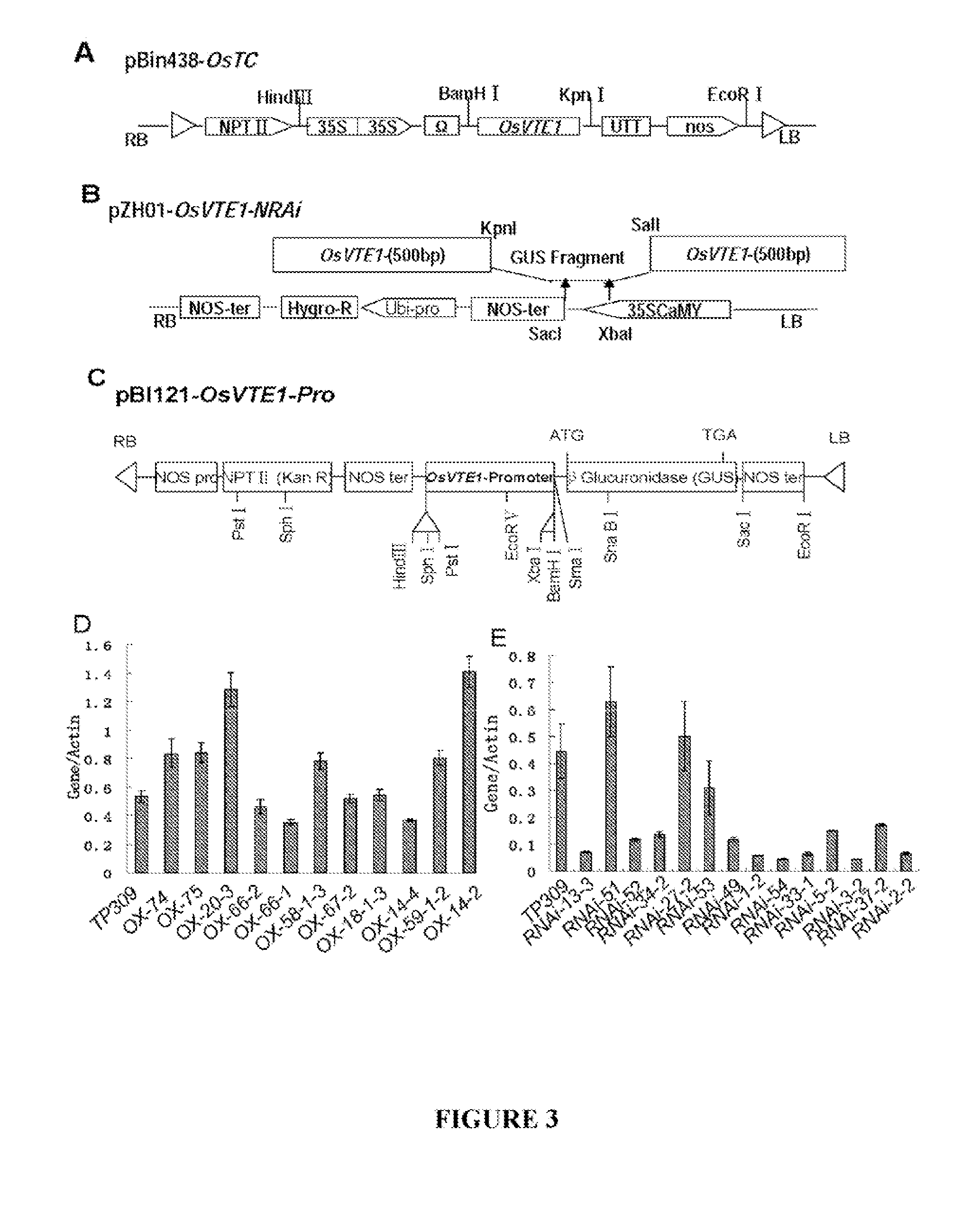
Genes Conferring Drought and Salt Tolerance and Uses Thereof
a technology of drought tolerance and genes, applied in the field of genes conferring drought tolerance, can solve the problems of reducing plant growth and agricultural productivity, and achieve the effects of increasing drought and salt tolerance to plants, reducing chlorophyll loss, and reducing h2o2 production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
RNA Isolation, RT-PCR and Real-Time Quantitative RT-PCR Analysis
[0111]The expression pattern of OsVTE1 (Os02g0276500) was investigated in rice seedlings (Oryza sativa, subsp. japonica cv. Taipei309; also referred to herein as TP309) using reverse transcription RT-PCR under various treatments. Total RNA isolation was performed following the method description by Zhang et al. (1999b). First-strand cDNA synthesis was primed with Oligo(dT)15 and catalyzed with M-MLV reverse transcriptase (Promega) at 37° C. for 1.5 hours. Reaction products were diluted 5-fold and used as templates for RT-PCR and real-time quantitative RT-PCR analysis.
[0112]Gene-specific primers, RT-OsVTE1 (5′-AGGGCCTATTCATCTCTACC-3′; SEQ ID NO: 4) and RT-OsVTE2 (5′-GGTGTCCATTCCCGAGTGCAGGCA-3′; SEQ ID NO: 5), were used for RT-PCR. Real-time quantitative RT-PCR analysis was performed using SYBR® Green PCR, an ABI PRISM® 7000 sequence detection system (Applied Biosystems), and gene-specific primers, Rtime1 (5′-TGCAATGTCTTC...
example 2
[0115]As shown in FIG. 3C, the OsVTE1 gene promoter (a 1.3 kB fragment upstream of the translation start site) was amplified using primers 5′-CCAAGCTTGCACGACCATAGG CGTGGGT-3′ (SEQ ID NO: 8) and 5′-GCTCTAGAGCTGATGCTGCGGGCGGGCA-3′ (SEQ ID NO: 9) and cloned into the HindI and BamHI sites of pBI121 containing a β-glucuronidase (GUS) reporter gene. The resulting construct was transferred into TP309 rice by Agrobacterium tumefaciens-mediated transformation as described by Hiei et al. (1994). GUS assays were performed at different developmental stages according to the method of Jefferson et al. (1987), and approximately fifteen independent T2 positive transgenic lines were used for the GUS staining assay shown in FIGS. 2C(a)-(h). Consistent with the results obtained from the RT-PCR assays, GUS was expressed in the stem, spikelet and leaf.
example 3
Construction of Overexpression and RNAi Vectors
[0116]As shown in FIG. 3A, the overexpression vector pBin438-OsTC was created by amplifying the full-length OsVTE1 (Os02g0276500) cDNA sequence using RT-PCR and gene-specified primer pairs 5′-CGGGGTACCAGGGCCTATTCATCTCTACC-3′ (SEQ ID NO: 10) and 5′-CGCGGATCCAGCATCAGCATGGACCTCGC-3′ (SEQ ID NO: 11). The full length sequence was cloned into the Kpn I and BamH I sites of the binary vector pBIN438 as described previously (Xie et al., 2002). Gene expression was driven by two copies of the 35S promoter, and the tobacco mosaic virus omega sequence was included downstream of the 35S promoter to enhance translation efficiency. The construct was introduced into Agrobacterium tumefaciens strain AGL1 and then transformed into TP309 rice as described by Hiei et al. (1994). A vector for overexpressing SEQ ID NO: 3 in corn is created using a similar protocol.
[0117]As shown in FIG. 3B, the RNAi vector pZH01-OsVTE1 was constructed by amplifying a 489-bp f...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Tm | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com