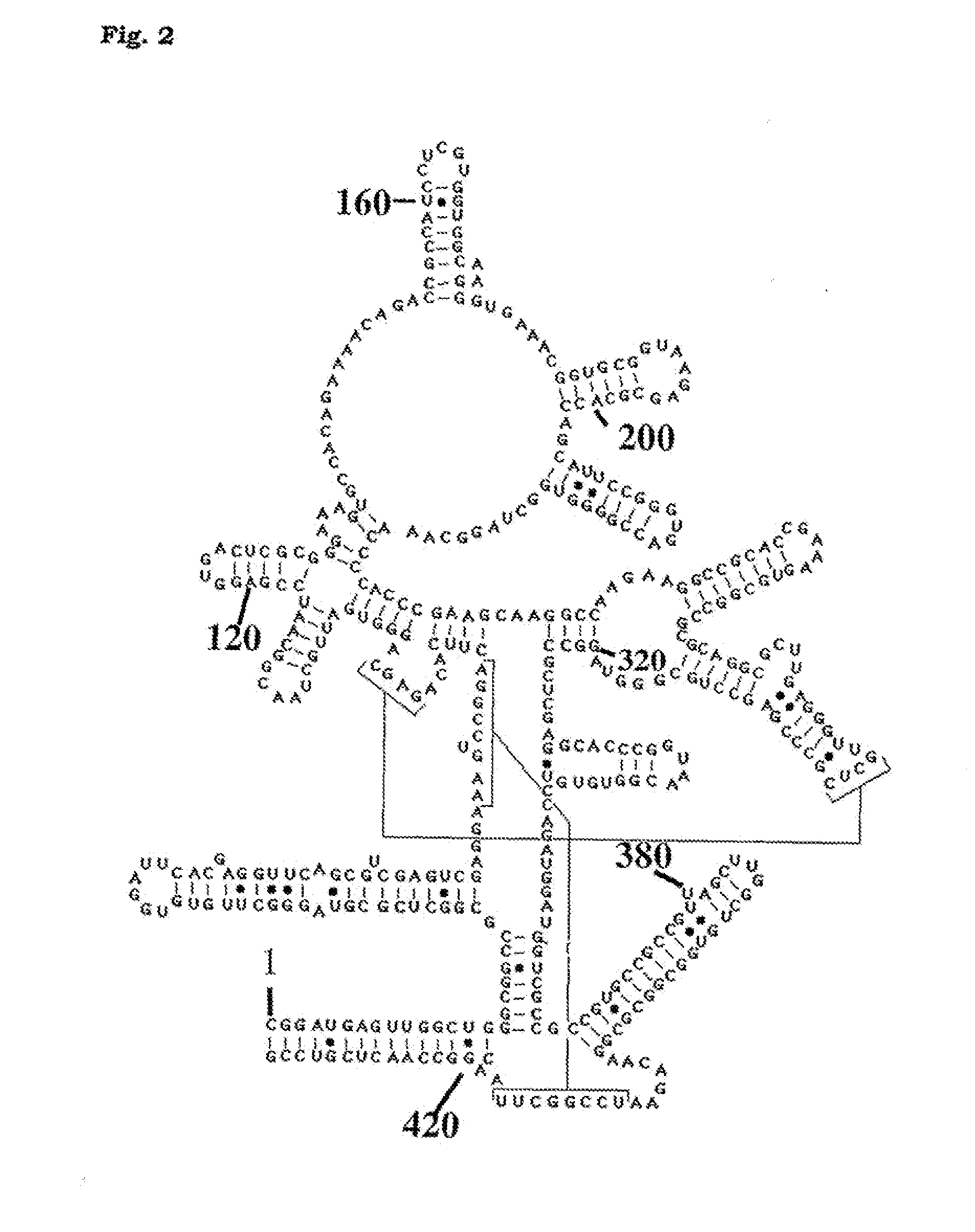
Method for detection of pathogenic organisms
a pathogenic organism and detection method technology, applied in the field of pathogenic organism detection, can solve the problems of time-consuming and difficult or impo differentiation between species of the same genus
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Mycobacteria
Materials and Methods
Bacterial Strains.
[0032]Mycobacteria used in this study are listed in Table 1 below. Clinical strains were already typed by 16S RNA gene sequencing in their respective institutes of provenance.
[0033]The RNase P RNA sequences for M. tuberculosis, M. bovis BCG and M. leprae were retrieved from the GenBank sequence database.
TABLE 1StrainSpeciesdescriptionProvenanceM. gastriATCC 15754ATCCM. kansasiiATCC 12748ATCCM. intracellulareD673Trudeau MycobacterialCollectionM. xenopiATCC 19276ATCCM. smegmatismc2 155Ref. 26M. aviumD702Trudeau MycobacterialCollectionM. marinumclinical strainResearch Centre BorstelM. fortuitumclinical strainResearch Centre BorstelM. malmoenseclinical strainResearch Centre BorstelM. paratuberculosis6783G-F Gerlach School ofVeterinary Medicine,Hanover GermanyM. gordoniaeclinical strainResearch Centre BorstelM. celatumclinical strainResearch Centre Borstel
PCR Amplification of the RNase P RNA Genes.
[0034]Based on the published sequences ...
example 2
Materials and Methods
Bacterial Strains.
[0061]DNA of analysed organisms were released by standard Proteinase K treatment of cell culture grown organisms and phenol extracted or were provided as purified DNA-preparations (Table 2).
TABLE 2Strains, host of origin, references, sources and accession numbersStrainHost of originReferenceSource*Accession no.6BCT (VR-125T)†PsittacineGolub & Wagner (1947)StoreyAJ012169GDDuckIllner (1960)NADCNJ1TurkeyPage (1959)NADCWCCattlePage (1967)NADCVS225PsittacineNADC360DuckStoreyN352DuckRichmond et al. (1982)StoreyCal10HumanFrancis & Magill (1938)StoreyCP3 (VR-574)PigeonPage & Bankowski (1960)NADCM56 (VR-630)MuskratSpalatio et al. (1966)NADCB577T (VR-656T)Ovine, abortionPerez-Martinez & Storz (1985)DenamurAJ131092EBABovine, abortionPerez-Martinez & Storz (1985)NADCOSPOvineAndersen & Van Deusen (1988)NADC EAE / LxOvine, abortionStoreyAB7Ovine, abortionRodolakis et al. (1989)RodolakisOC1Ovine, conjunctivitisRodolakis et al. (1989)RodolakisAV1Bovine...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com