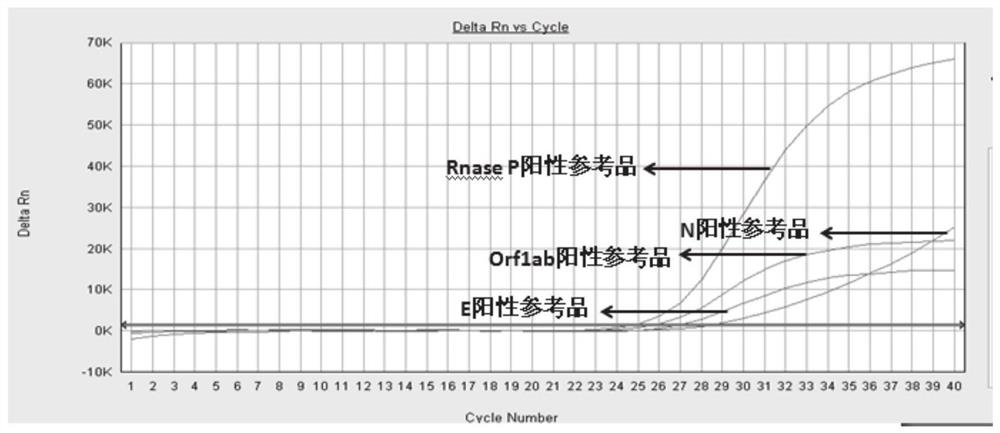
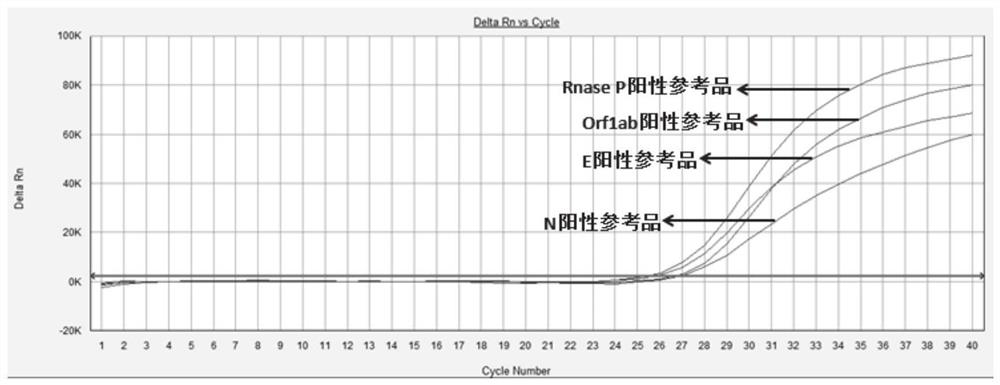
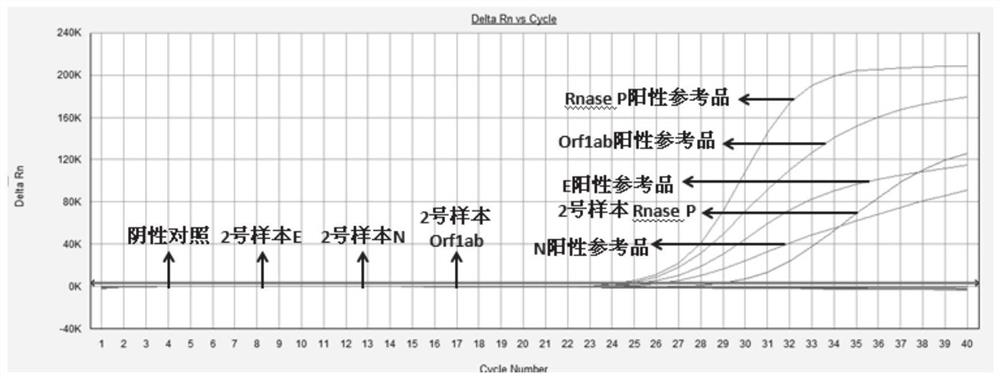
Primers and method for simultaneously detecting Orf1ab, E and N regions of 2019-nCov virus in one tube
A 2019-ncov and detection method technology, applied in the fields of life sciences and biology, can solve the problems of missed diagnosis, low detection accuracy and false positives of infected people in the incubation period, so as to improve specificity and accuracy, save detection cost and detection time Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Real-time fluorescence quantitative PCR kit for detecting 2019-nCov or SARS-Cov-2 virus nucleic acid, mainly including:
[0052] (1) Viral RNA extraction reagent;
[0053] (2) Reagents for reverse transcription and PCR amplification: one-step reverse transcription-fluorescent quantitative reagents;
[0054] (3) Specific primers and Taqman fluorescent probes for amplification, including upstream and downstream primers and probes for amplifying the three regions of Orf1ab, E and N covering the 2019-nCov virus nucleic acid, Orf1ab-F, Orf1ab respectively -R, Orf1ab-Probe; E-F, E-R, E-Probe; N-F, N-R, N-Probe; the upstream and downstream primers and probes used to detect the internal standard gene Rnase P are Rnase P-F, Rnase P-R, and Rnase P-Probe respectively; The base sequence is:
[0055] orflab-F: 5'-TGAGTGAAATGGTCATGTGTGG-3';
[0056] orflab-Pro: 5'-FAM-CAGGTGGAACCTCATCAGGAGATGC-BHQ1-3';
[0057] orflab-R: 5'-ACACTATTAGCATAAGCAGTTGTGG-3';
[0058] E gene-F: 5'-CGG...
Embodiment 2
[0074] The operation process of the inventive method:
[0075] 1. Extract viral RNA from throat swabs (serum, plasma, etc.):
[0076] (1) Use a pipette to add 560 μL of Carrier RNA working solution (the ratio of Lysis Buffer RL to Carrier RNA is 100:1) into a 1.5 mL centrifuge tube.
[0077] (2) Add 140 μL throat swab culture solution or serum, plasma and other specimens to the centrifuge tube, vortex for 15 sec to mix, and incubate at room temperature (15-25°C) for 10 min.
[0078] (3) Add 560 μL of absolute ethanol after centrifugation, and vortex for 15 sec (when the ambient temperature is higher than 25°C, the absolute ethanol needs to be pre-cooled in advance).
[0079] (4) After the instant centrifugation, transfer 630 μL to the adsorption column CR2, centrifuge at 8000 rpm for 1 min, discard the waste liquid, and put the adsorption column back into the collection tube.
[0080] (5) Pass the remaining liquid in the centrifuge tube through the column again according to ...
Embodiment 3
[0092] Embodiment 3 Sensitivity detection of this kit
[0093] With the plasmid concentration being 1000, 100, 10copies / μL as a template, the experiment was carried out according to the method shown in Example 2, each concentration was repeated 10 times, and a blank control was set at the same time. As a result, it was found that the detection limit of the present invention was 1000 copies / μL.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com