Distinguishing methylation levels in complex biological samples
a biological sample and complex technology, applied in the field of methylation pattern determination in genomic dna, can solve the problems of compromising the accuracy and affordability of currently available techniques, affecting the development of methylation detection techniques into robust and cost efficient screening tools, and often using cumbersome and expensive purification techniques to purify genomic samples
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example i
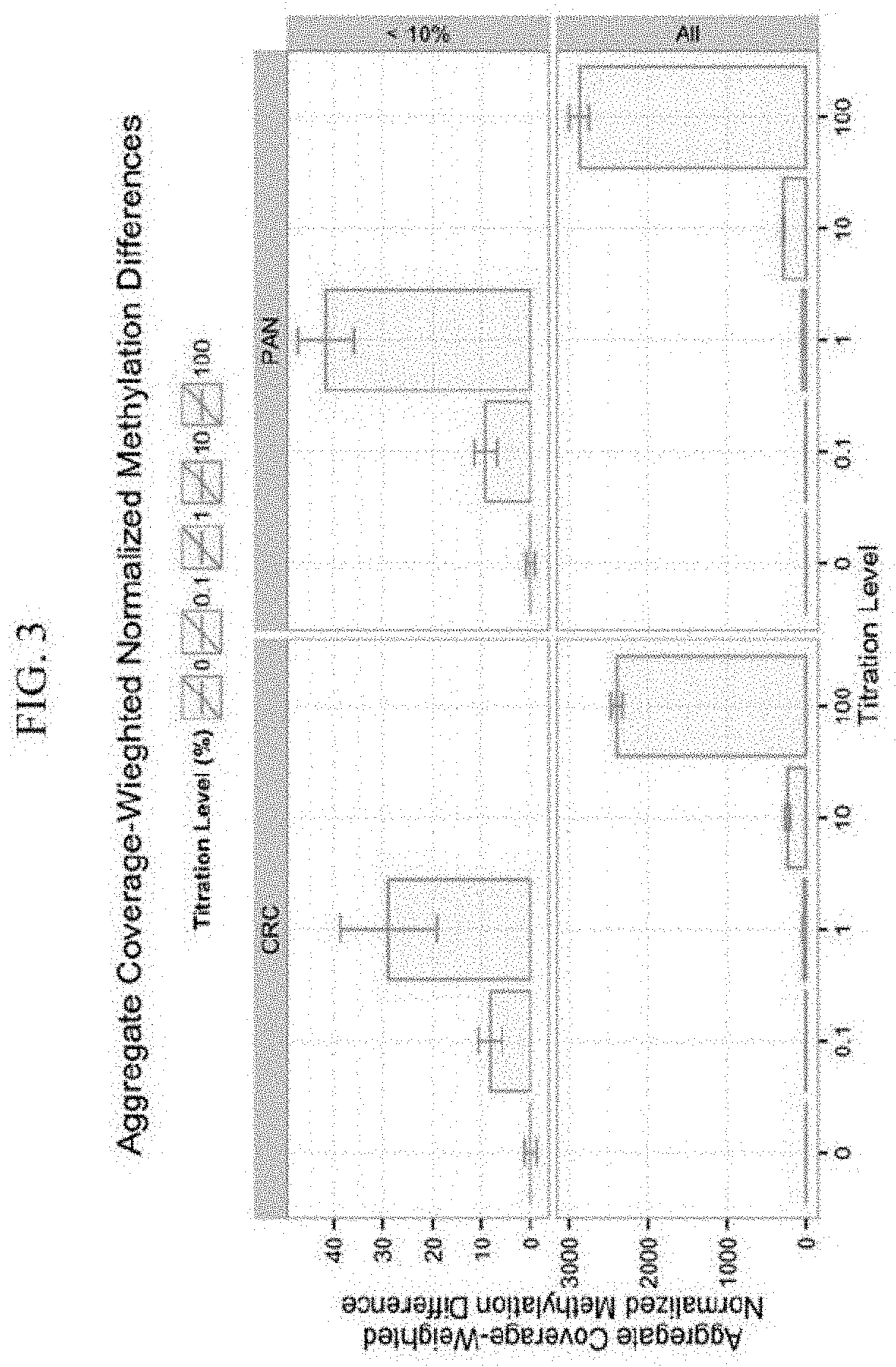
Analytical Sensitivity of ctDNA Methylation-Based Cancer Detection Using Aggregate Normalized Coverage-Weighted Methylation Differences
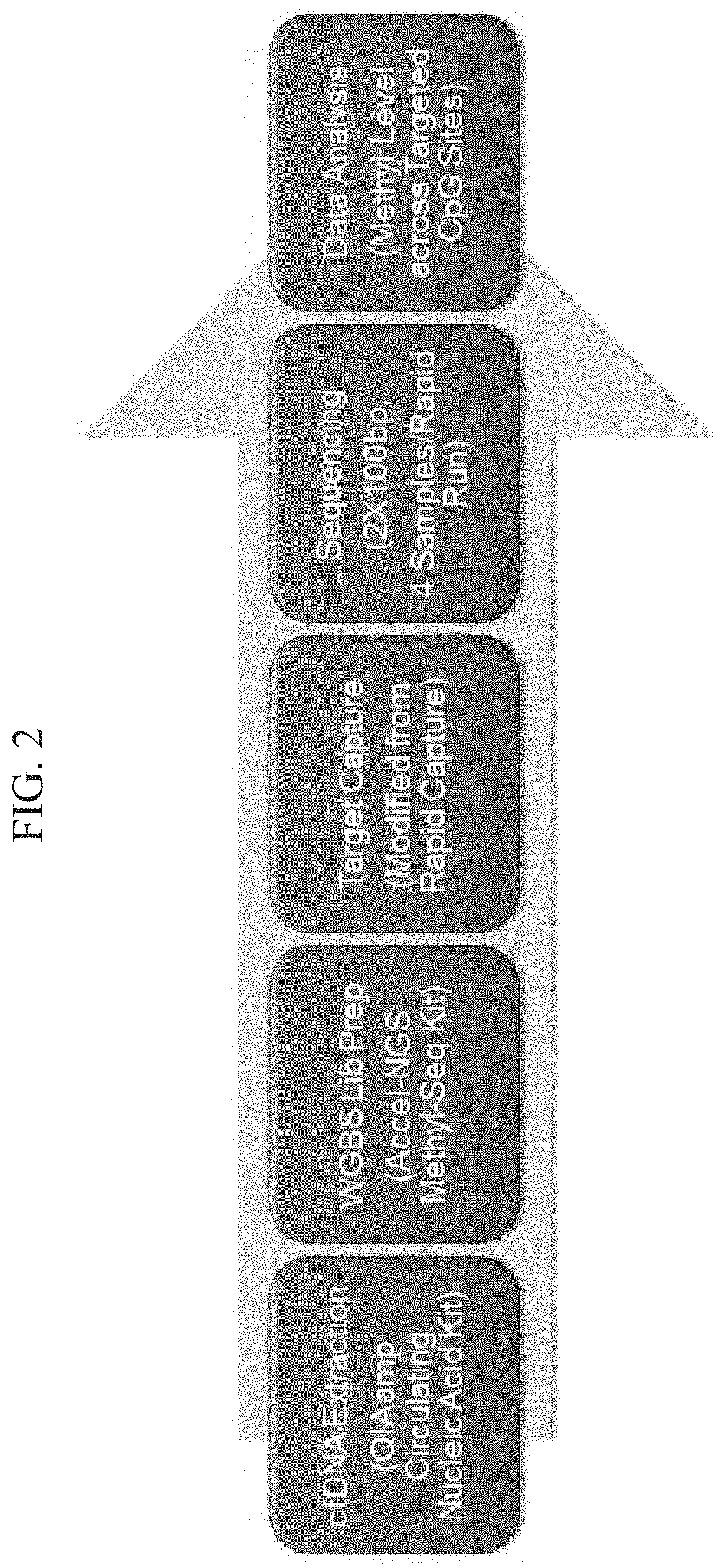
[0154]This example describes a highly sensitive assay for detecting methylation in circulating tumor DNA (ctDNA). Aberrant DNA methylation is a widespread phenomenon in cancer and may be among the earliest changes to occur during oncogenesis. The assay described in this example can be useful for cancer screening.
[0155]The general approach applied here includes targeted methylation sequencing for multiple CpG sites affected in cancer.
[0156]Technical challenges addressed by the approach include providing ultra-high sensitivity and specificity that benefits screening applications, providing a protocol for targeted methyl-seq from low input ctDNA, and providing bioinformatics algorithms for analysis of methylation levels across a large number of targeted sites.
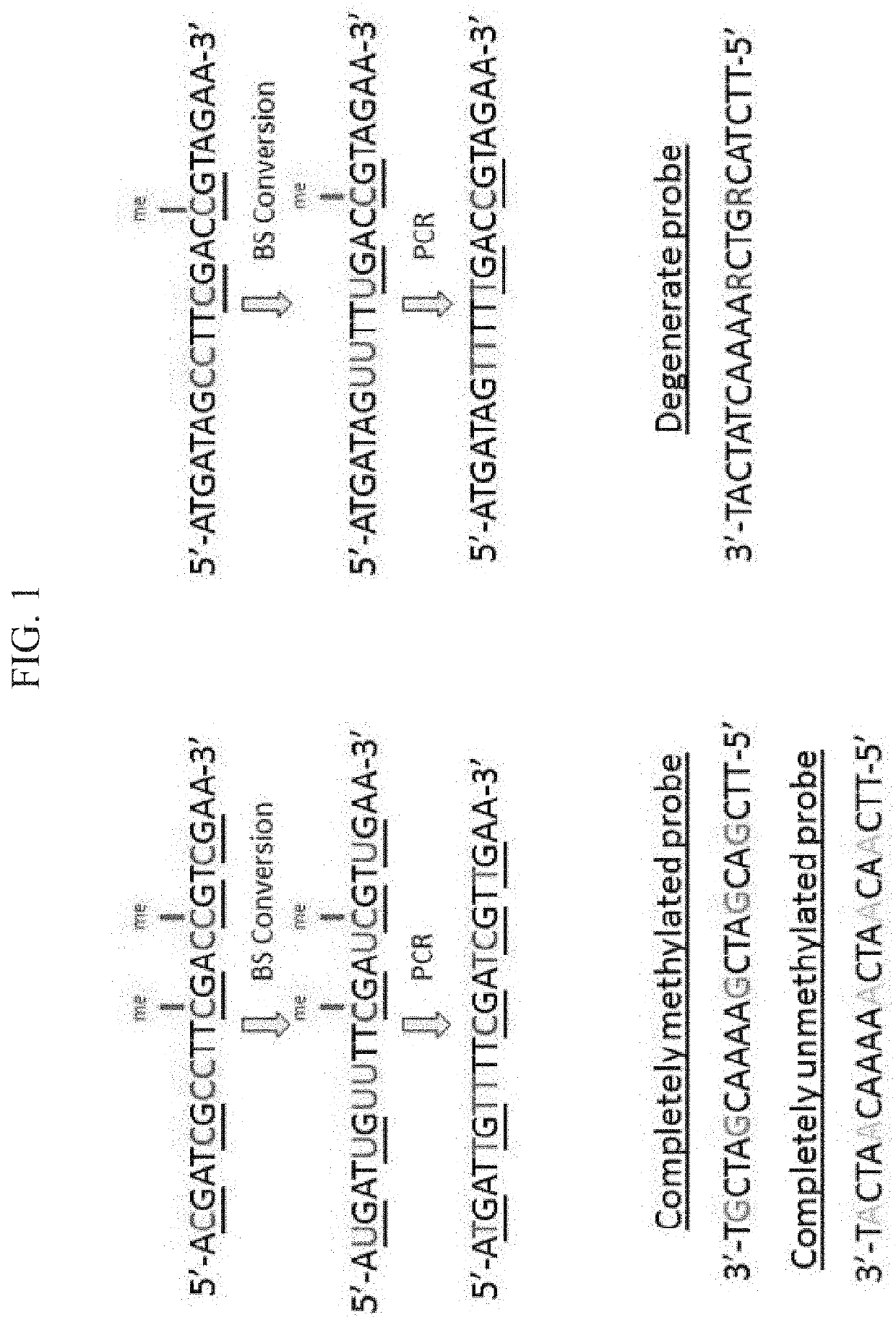
Targeted Capture Probe Design
[0157]Two targeted methylation panels were developed. The Pan-Canc...
example ii
Analytical Sensitivity of ctDNA Methylation-Based Cancer Detection Using Coverage-Weighted Methylation Scores
[0171]This example describes an alternative highly sensitive assay for detecting methylation in circulating tumor DNA (ctDNA). The assay described in this example also can be useful for cancer screening, monitoring disease progression, or evaluating a patient's response to a therapeutic treatment.
Targeted Capture Probe Design
[0172]For this study, the two targeted methylation panels described in Example I were pooled together. The Pan-Cancer Panel targets 9,921 affected CpG sites in 20 major cancer types as selected from The Cancer Genome Atlas Database. The CpG sites included in the Pan-Cancer Panel are listed in Table I. The CRC Panel targets 1,162 affected CpG sites in colorectal cancer. The CpG sites included in the CRC Panel are listed in Table II. The combined CpG sites listed in Table I and Table II refer to Genome Build 37.
[0173]The probe sequences for the CpG sites we...
example iii
Clinical Performance of ctDNA Methylation-Based Cancer Detection Using Normalized Coverage-Weighted Methylation Score Differences
[0187]This example evaluates clinical sensitivity and specificity of the methylation-based cancer detection in circulating tumor DNA (ctDNA) using normalized coverage weighted methylation score differences. As noted above, the assay described in this example can be useful for cancer screening, monitoring disease progression, or evaluating a patient's response to a therapeutic treatment.
Targeted Capture Probe Design
[0188]For this study, the two targeted methylation panels described in Example I were pooled together. The Pan-Cancer Panel targets 9,921 affected CpG sites in 20 major cancer types as selected from The Cancer Genome Atlas Database. The CpG sites included in the Pan-Cancer Panel are listed in Table I. The CRC Panel targets 1,162 affected CpG sites in colorectal cancer. The CpG sites included in the CRC Panel are listed in Table II. The combined C...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com