Primer and method for quickly detecting IT15 gene CAG repeat sequence dynamic mutation
A repeat sequence and gene technology, applied in the field of biomedicine, can solve the problems of high experimental conditions, difficulty in popularizing and popularizing gene mutation detection technology, and difficulty in clinical application in general hospitals.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
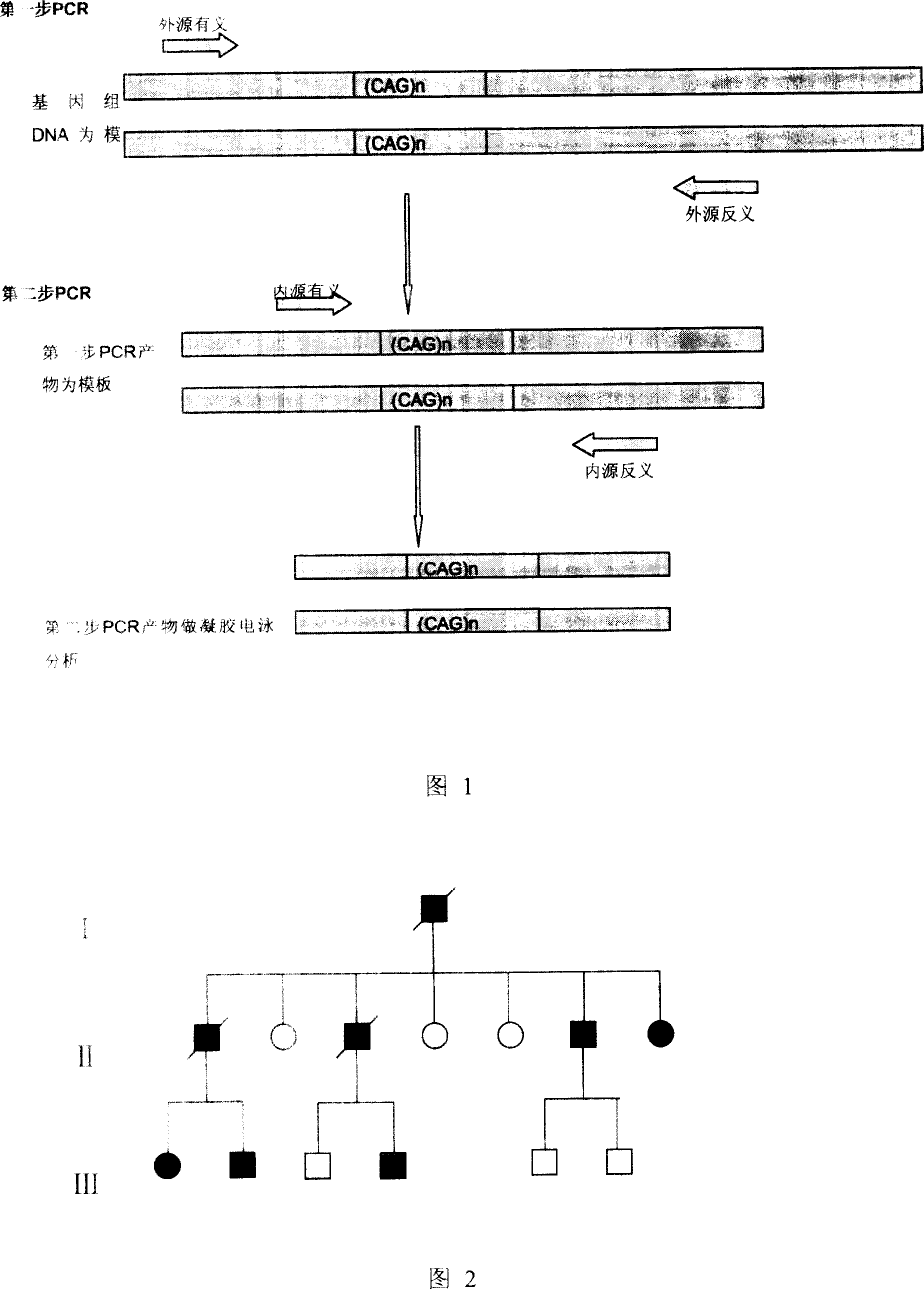
[0076] Example 1: A primer for rapidly detecting the dynamic mutation of the CAG repeat sequence of the IT15 gene, consisting of a first pair of primers corresponding to the first step of PCR amplification and a second pair of primers corresponding to the second step of PCR amplification;
[0077] The first pair of primers adopts any one pair of primers in the following four pairs of primers:
[0078] (I), exogenous sense: 5'CTTGCTGTGTGAGGCAGAACCTG-3'
[0079] Exogenous antisense: 5'GGCTGAGGAAGCTGAGGAG-3'
[0080] (II), exogenous sense: 5'TTCACACACAGCTTCGC-3'
[0081] Exogenous antisense: 5'CCTCCTCTCGAAGTGTCCCG-3'
[0082] (III), exogenous sense: 5'GGCAGAGTCCGCAGGCTAG-3'
[0083] Exogenous antisense: 5'AAGTTCCATAGCGATGCCCAG-3'
[0084] (IV), exogenous sense: 5'CTAGGGCTGTCAATCATGCTGG-3'
[0085] Exogenous antisense: 5'AAGTTCCATAGCGATGCCCAG-3'
[0086] The second pair of primers adopts any pair of primers in the following two pairs of primers:
[0087] (1), endogenous sen...
Embodiment 2
[0092] Embodiment 2: A method for rapidly detecting the dynamic mutation of the CAG repeat sequence of the IT15 gene, which is characterized in that it comprises the following steps:
[0093] Step 1: Prepare DNA
[0094] (1) Take blood samples from the human body;
[0095] (2) Obtaining DNA from blood samples, i.e. blood sample genomic DNA sample preparation:
[0096] Reagent preparation:
[0097] Anticoagulant: 0.48g citric acid, 1.32g sodium citrate, 1.47g glucose per 100ml;
[0098] Red blood cell lysate: 10mmol / L Tris-HCl, pH7.6;
[0099] 5mmol / L MgCl 2 ;
[0100] 10mmol / L NaCl;
[0101] White blood cell lysate: 10mmol / L Tris-HCl, pH7.6;
[0102] 10mmol / LEDTAPH8.0
[0103] 50mmol / L NaCl
[0104] 10mg / ml proteinase K (Protease K): 10mg Protease K dissolved in 1ml ddH 2 O. Aliquot and store at -20°C. When in use, melt at 4°C.
[0105] Procedure for obtaining DNA from a blood sample:
[0106] ①. Take 3ml of fresh blood and add 0.5ml of anticoagulant, mix well, c...
Embodiment 3
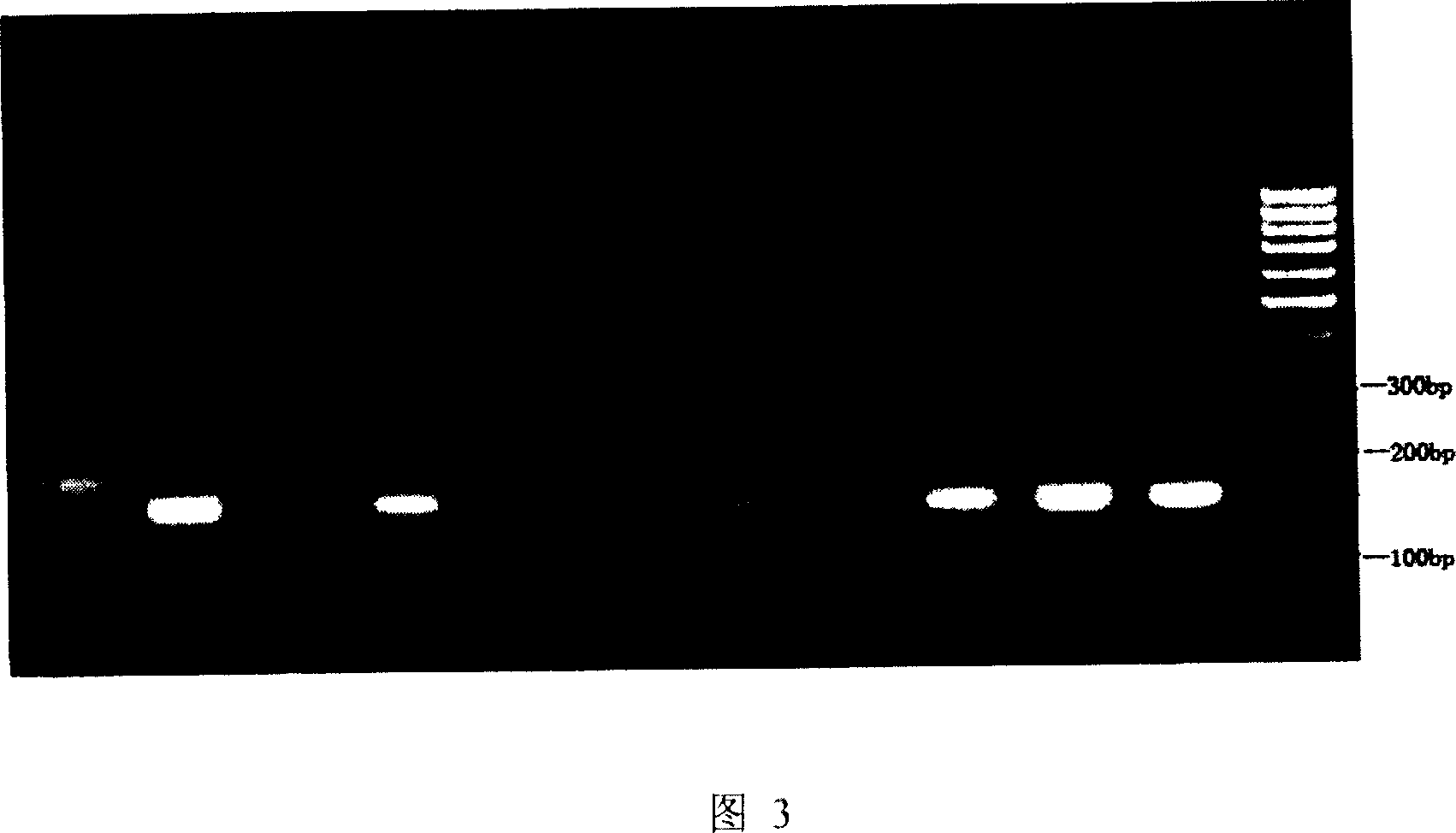
[0150] Example 3: Using the method of the present invention to rapidly detect the dynamic mutation of the IT15 gene CAG repeat sequence in a Huntington's disease family in Anhui Province.
[0151] 1. Materials and methods
[0152] 1.1 Research objects: A HD family in Anhui Province includes 4 generations of members, 11 of which were sampled and analyzed. 3ml of each peripheral venous blood (anticoagulated with 3.8% sodium citrate). The red blood cells were lysed, and the white blood cells were obtained by centrifugation, and the genomic DNA in the white blood cells was extracted by conventional saturated sodium chloride method.
[0153] 1.2 Amplify part of IT15 gene containing CAG repeat sequence. Using the first-step PCR primer (I) and the second-step PCR primer (I) mentioned in this patent, the htt gene of a chorea family member in Anhui Province was amplified according to the method in Example 2.
[0154] Amplification was performed using NEST-PCR. 1st PCR: The amplific...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com