Method for detecting polyploidy plant gene mononucleotide site mutation
A single nucleotide and point mutation technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of false positive detection, low efficiency, time-consuming, etc., and achieve good detection of point mutations, easy operation, The effect of easy method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
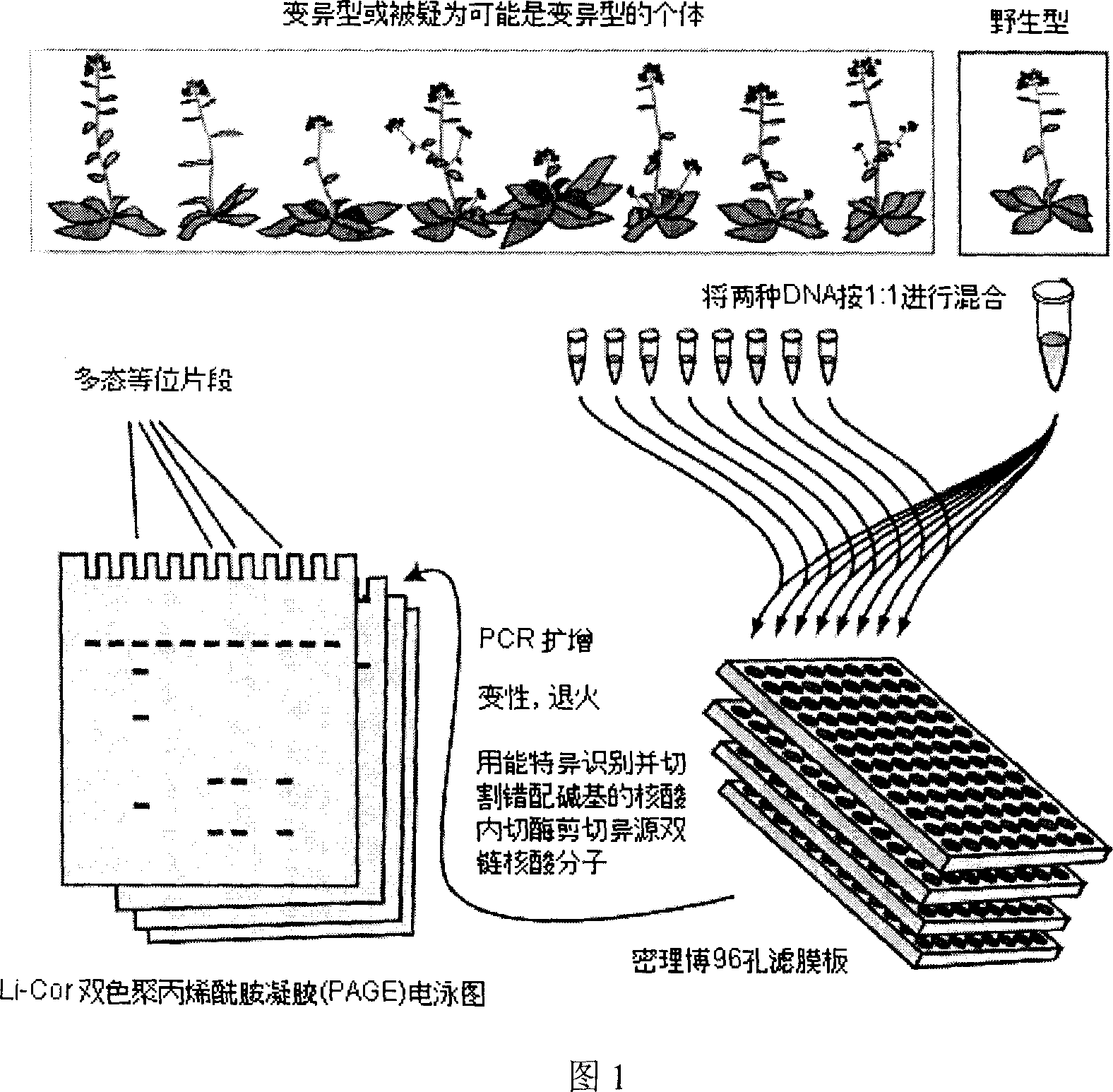
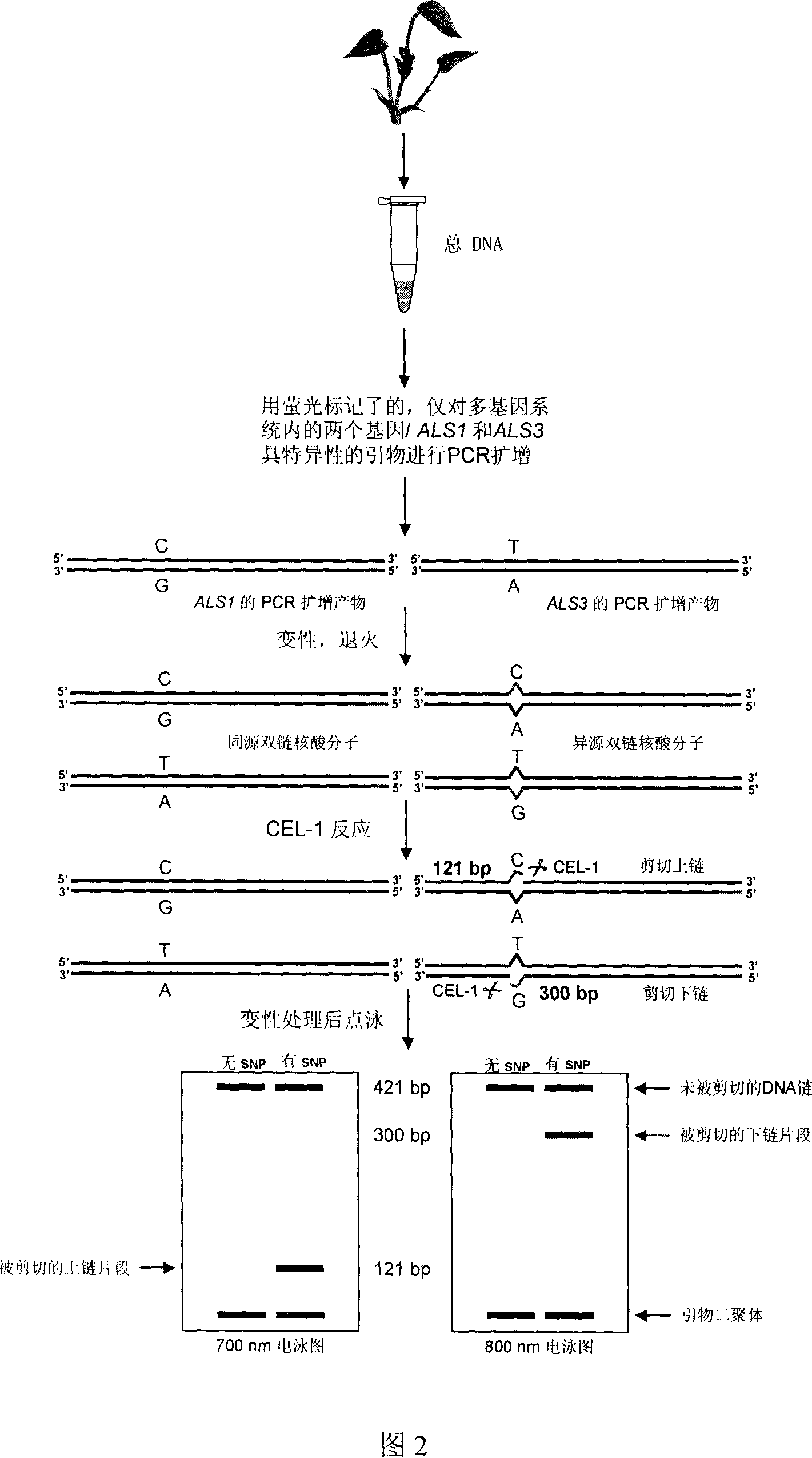
[0021] A method for detecting single nucleotide point mutations in polyploid plant genes, the specific steps of which are as follows:
[0022] 1. The material used is a paddy field weed with a multi-gene system. DNA extraction kit (QIAGEN-DNeasy Plant Mini Kit) was used to extract total DNA from a polyploid weed with a polygenic system: Cosmos miltiorrhiza;
[0023] 2. Design its multi-gene system (ALS1, ALS2, ALS3 and ALS4, according to Wang et al., Discovery of single-nucleotide mutations in acetolactate synthase genes by EcoTILLING, Pestic.Biochem.Physiol.(2006), doi: 10.1016 / j A pair (two) primers (the sequences of which are respectively:
[0024] TCTGCATCGCCACCTCG and TGAAGCAGATGGAGG GCAGG) for FirstPCR amplification (the two primers were labeled with 700nm and 800nm fluorescent dyes, respectively, and the labeling sequences were gctacggactgacctcggac and ctgacgtgatgctcctgacg). That is, the sequence of the forward primer is: gctacggactgacctcggacTCTGCATCGCCACCTCG, and t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com